Single-cell transcriptional profile of CD34+ hematopoietic progenitor cells from del(5q) myelodysplastic syndromes and impact of lenalidomide
- PMID: 38902243
- PMCID: PMC11189937
- DOI: 10.1038/s41467-024-49529-x
Single-cell transcriptional profile of CD34+ hematopoietic progenitor cells from del(5q) myelodysplastic syndromes and impact of lenalidomide
Abstract
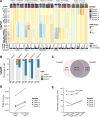
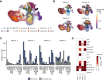
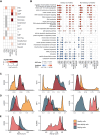
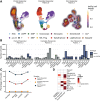
While myelodysplastic syndromes with del(5q) (del(5q) MDS) comprises a well-defined hematological subgroup, the molecular basis underlying its origin remains unknown. Using single cell RNA-seq (scRNA-seq) on CD34+ progenitors from del(5q) MDS patients, we have identified cells harboring the deletion, characterizing the transcriptional impact of this genetic insult on disease pathogenesis and treatment response. Interestingly, both del(5q) and non-del(5q) cells present similar transcriptional lesions, indicating that all cells, and not only those harboring the deletion, may contribute to aberrant hematopoietic differentiation. However, gene regulatory network (GRN) analyses reveal a group of regulons showing aberrant activity that could trigger altered hematopoiesis exclusively in del(5q) cells, pointing to a more prominent role of these cells in disease phenotype. In del(5q) MDS patients achieving hematological response upon lenalidomide treatment, the drug reverts several transcriptional alterations in both del(5q) and non-del(5q) cells, but other lesions remain, which may be responsible for potential future relapses. Moreover, lack of hematological response is associated with the inability of lenalidomide to reverse transcriptional alterations. Collectively, this study reveals transcriptional alterations that could contribute to the pathogenesis and treatment response of del(5q) MDS.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

