PatchSorter: a high throughput deep learning digital pathology tool for object labeling
- PMID: 38902336
- PMCID: PMC11190251
- DOI: 10.1038/s41746-024-01150-4
PatchSorter: a high throughput deep learning digital pathology tool for object labeling
Abstract
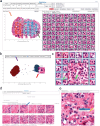
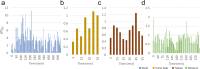
The discovery of patterns associated with diagnosis, prognosis, and therapy response in digital pathology images often requires intractable labeling of large quantities of histological objects. Here we release an open-source labeling tool, PatchSorter, which integrates deep learning with an intuitive web interface. Using >100,000 objects, we demonstrate a >7x improvement in labels per second over unaided labeling, with minimal impact on labeling accuracy, thus enabling high-throughput labeling of large datasets.
© 2024. The Author(s).
Conflict of interest statement
A.M. is an equity holder in Picture Health, Elucid Bioimaging, and Inspirata Inc. Currently, he serves on the advisory board of Picture Health, Aiforia Inc., and SimBioSys. He also currently consults for SimBioSys. He also has sponsored research agreements with AstraZeneca, Boehringer-Ingelheim, Eli-Lilly, and Bristol Myers-Squibb. His technology has been licensed to Picture Health and Elucid Bioimaging. He is also involved in three different R01 grants with Inspirata Inc. L.B. is a consultant for Sangamo and Protalix and is on the scientific advisory boards of Vertex and Nephcure. A.J. provides consulting for Merck, Lunaphore, and Roche, the latter of which he also has a sponsored research agreement. H.M.H. received financial compensation from Roche Diagnostics BV paid to the institute. No other conflicts of interest were declared.
Figures
References
-
- Bengar, J. Z., van de Weijer, J., Twardowski, B. & Raducanu, B. Reducing label effort: self-supervised meets active learning. In Proc. IEEE/CVF International Conference on Computer Vision 1631–1639 (IEEE, 2021).
Grants and funding
- U24 DK100845/DK/NIDDK NIH HHS/United States
- U01 DK100867/DK/NIDDK NIH HHS/United States
- U2C TR002818/TR/NCATS NIH HHS/United States
- U2CTR002818/U.S. Department of Health & Human Services | NIH | National Center for Advancing Translational Sciences (NCATS)
- R01 LM013864/LM/NLM NIH HHS/United States
- U01 DK133090/DK/NIDDK NIH HHS/United States
- U54 DK083912/DK/NIDDK NIH HHS/United States
- UM1 DK100846/DK/NIDDK NIH HHS/United States
- U01 CA248226/CA/NCI NIH HHS/United States
- U01 DK100876/DK/NIDDK NIH HHS/United States
- UM1 DK100845/DK/NIDDK NIH HHS/United States
- UM1 DK100866/DK/NIDDK NIH HHS/United States
- 2R01DK118431-04/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- 1R01LM013864/U.S. Department of Health & Human Services | NIH | U.S. National Library of Medicine (NLM)
- 1U01DK133090/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01 DK118431/DK/NIDDK NIH HHS/United States
- U01 DK100866/DK/NIDDK NIH HHS/United States
- U01 DK100846/DK/NIDDK NIH HHS/United States
- UM1 DK100876/DK/NIDDK NIH HHS/United States
- UM1 DK100867/DK/NIDDK NIH HHS/United States
- U54DK083912/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- U01 CA239055/CA/NCI NIH HHS/United States

