CREB1 promotes expression of immune checkpoint HLA-E leading to immune escape in multiple myeloma
- PMID: 38902472
- PMCID: PMC11286514
- DOI: 10.1038/s41375-024-02303-w
CREB1 promotes expression of immune checkpoint HLA-E leading to immune escape in multiple myeloma
Abstract
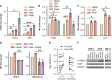
Multiple myeloma (MM) cells effectively escape anti-tumoral immunity to survive in the tumor microenvironment (TME). Herein, we identify non-classical major histocompatibility complex (MHC) class I molecule HLA-E as a major contributing factor in immune escape. Clinically, HLA-E expression correlates with aggressive disease features such as t(4;14) and CD56 expression and is induced by IFN-gamma (IFN-γ) in the TME. We discovered that HLA-E is regulated by cAMP responsive element binding protein 1 (CREB1) transcription factor by direct promoter binding; genomic and pharmacological inhibition of CREB1 reduced HLA-E levels even in the presence of IFN-γ or IFN-γ activating agents, such as immunomodulatory drugs and panobinostat. HLA-E binds to natural killer group 2A (NKG2A), delivering an inhibitor signal to natural killer (NK) cells. Treatment with a CREB1 inhibitor was able to restore NK cell-mediated cytotoxicity against MM cell lines and patient samples. In conclusion, our results strongly demonstrate that CREB1 inhibition promotes anti-tumoral immunity in MM by limiting HLA-E expression and enhancing the activity of NK cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

