Non-stem cell lineages as an alternative origin of intestinal tumorigenesis in the context of inflammation
- PMID: 38902475
- PMCID: PMC11250264
- DOI: 10.1038/s41588-024-01801-y
Non-stem cell lineages as an alternative origin of intestinal tumorigenesis in the context of inflammation
Abstract
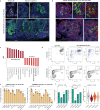
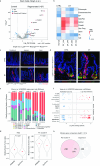
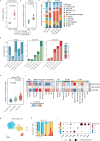
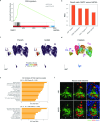
According to conventional views, colon cancer originates from stem cells. However, inflammation, a key risk factor for colon cancer, has been shown to suppress intestinal stemness. Here, we used Paneth cells as a model to assess the capacity of differentiated lineages to trigger tumorigenesis in the context of inflammation in mice. Upon inflammation, Paneth cell-specific Apc mutations led to intestinal tumors reminiscent not only of those arising in patients with inflammatory bowel disease, but also of a larger fraction of human sporadic colon cancers. The latter is possibly because of the inflammatory consequences of western-style dietary habits, a major colon cancer risk factor. Machine learning methods designed to predict the cell-of-origin of cancer from patient-derived tumor samples confirmed that, in a substantial fraction of sporadic cases, the origins of colon cancer reside in secretory lineages and not in stem cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Update of
-
Paneth cells as the origin of intestinal cancer in the context of inflammation.Res Sq [Preprint]. 2023 Jan 19:rs.3.rs-2458794. doi: 10.21203/rs.3.rs-2458794/v1. Res Sq. 2023. Update in: Nat Genet. 2024 Jul;56(7):1456-1467. doi: 10.1038/s41588-024-01801-y. PMID: 36711533 Free PMC article. Updated. Preprint.
References
-
- Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. - PubMed
-
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. - PubMed
-
- Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. - PubMed
MeSH terms
Substances
Grants and funding
- P30-013330/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01CA229216/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- T32 AG023475/AG/NIA NIH HHS/United States
- 2014-1181/World Cancer Research Fund (WCRF UK)
- R01 CA229216/CA/NCI NIH HHS/United States
- R01 CA214625/CA/NCI NIH HHS/United States
- 220002/WT_/Wellcome Trust/United Kingdom
- P30 AG038072/AG/NIA NIH HHS/United States
- 11407/KWF Kankerbestrijding (Dutch Cancer Society)
- 2018-2025 312041/Academy of Finland (Suomen Akatemia)
- P30-AG038072/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

