A single-center, retrospective study of hospitalized patients with lower respiratory tract infections: clinical assessment of metagenomic next-generation sequencing and identification of risk factors in patients
- PMID: 38902783
- PMCID: PMC11191188
- DOI: 10.1186/s12931-024-02887-y
A single-center, retrospective study of hospitalized patients with lower respiratory tract infections: clinical assessment of metagenomic next-generation sequencing and identification of risk factors in patients
Abstract
Introduction: Lower respiratory tract infections(LRTIs) in adults are complicated by diverse pathogens that challenge traditional detection methods, which are often slow and insensitive. Metagenomic next-generation sequencing (mNGS) offers a comprehensive, high-throughput, and unbiased approach to pathogen identification. This retrospective study evaluates the diagnostic efficacy of mNGS compared to conventional microbiological testing (CMT) in LRTIs, aiming to enhance detection accuracy and enable early clinical prediction.
Methods: In our retrospective single-center analysis, 451 patients with suspected LRTIs underwent mNGS testing from July 2020 to July 2023. We assessed the pathogen spectrum and compared the diagnostic efficacy of mNGS to CMT, with clinical comprehensive diagnosis serving as the reference standard. The study analyzed mNGS performance in lung tissue biopsies and bronchoalveolar lavage fluid (BALF) from cases suspected of lung infection. Patients were stratified into two groups based on clinical outcomes (improvement or mortality), and we compared clinical data and conventional laboratory indices between groups. A predictive model and nomogram for the prognosis of LRTIs were constructed using univariate followed by multivariate logistic regression, with model predictive accuracy evaluated by the area under the ROC curve (AUC).
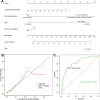
Results: (1) Comparative Analysis of mNGS versus CMT: In a comprehensive analysis of 510 specimens, where 59 cases were concurrently collected from lung tissue biopsies and BALF, the study highlights the diagnostic superiority of mNGS over CMT. Specifically, mNGS demonstrated significantly higher sensitivity and specificity in BALF samples (82.86% vs. 44.42% and 52.00% vs. 21.05%, respectively, p < 0.001) alongside greater positive and negative predictive values (96.71% vs. 79.55% and 15.12% vs. 5.19%, respectively, p < 0.01). Additionally, when comparing simultaneous testing of lung tissue biopsies and BALF, mNGS showed enhanced sensitivity in BALF (84.21% vs. 57.41%), whereas lung tissues offered higher specificity (80.00% vs. 50.00%). (2) Analysis of Infectious Species in Patients from This Study: The study also notes a concerning incidence of lung abscesses and identifies Epstein-Barr virus (EBV), Fusobacterium nucleatum, Mycoplasma pneumoniae, Chlamydia psittaci, and Haemophilus influenzae as the most common pathogens, with Klebsiella pneumoniae emerging as the predominant bacterial culprit. Among herpes viruses, EBV and herpes virus 7 (HHV-7) were most frequently detected, with HHV-7 more prevalent in immunocompromised individuals. (3) Risk Factors for Adverse Prognosis and a Mortality Risk Prediction Model in Patients with LRTIs: We identified key risk factors for poor prognosis in lower respiratory tract infection patients, with significant findings including delayed time to mNGS testing, low lymphocyte percentage, presence of chronic lung disease, multiple comorbidities, false-negative CMT results, and positive herpesvirus affecting patient outcomes. We also developed a nomogram model with good consistency and high accuracy (AUC of 0.825) for predicting mortality risk in these patients, offering a valuable clinical tool for assessing prognosis.
Conclusion: The study underscores mNGS as a superior tool for lower respiratory tract infection diagnosis, exhibiting higher sensitivity and specificity than traditional methods.
Keywords: Diagnostic efficacy; Lower respiratory tract infections (LRTIs); Metagenomic next-generation sequencing (mNGS); Nomogram; Predictive model.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Clinical utility of metagenomic next-generation sequencing in pathogen detection for lower respiratory tract infections.Sci Rep. 2025 May 30;15(1):19039. doi: 10.1038/s41598-025-03564-w. Sci Rep. 2025. PMID: 40447717 Free PMC article.
-
Diagnostic value of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in patients with lower respiratory tract infections.Diagn Microbiol Infect Dis. 2025 Feb;111(2):116620. doi: 10.1016/j.diagmicrobio.2024.116620. Epub 2024 Nov 17. Diagn Microbiol Infect Dis. 2025. PMID: 39586148
-
Clinical utility of metagenomic next-generation sequencing on bronchoalveolar lavage fluid in diagnosis of lower respiratory tract infections.BMC Pulm Med. 2024 Aug 29;24(1):422. doi: 10.1186/s12890-024-03237-w. BMC Pulm Med. 2024. PMID: 39210307 Free PMC article.
-
Metagenomic next generation sequencing of bronchoalveolar lavage fluids for the identification of pathogens in patients with pulmonary infection: A retrospective study.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116402. doi: 10.1016/j.diagmicrobio.2024.116402. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38878340 Review.
-
Metagenomic next-generation sequencing on treatment strategies and prognosis of patients with lower respiratory tract infections: A systematic review and meta-analysis.Int J Antimicrob Agents. 2025 Mar;65(3):107440. doi: 10.1016/j.ijantimicag.2024.107440. Epub 2025 Jan 4. Int J Antimicrob Agents. 2025. PMID: 39761759
Cited by
-
Clinical utility of metagenomic next-generation sequencing in pathogen detection for lower respiratory tract infections.Sci Rep. 2025 May 30;15(1):19039. doi: 10.1038/s41598-025-03564-w. Sci Rep. 2025. PMID: 40447717 Free PMC article.
-
Biomarkers (NLR, PLR, SII) for Frequent COPD Exacerbations: Diagnostic and Clinical Management Implications in a Retrospective Study.Int J Chron Obstruct Pulmon Dis. 2025 Apr 5;20:987-998. doi: 10.2147/COPD.S510118. eCollection 2025. Int J Chron Obstruct Pulmon Dis. 2025. PMID: 40207023 Free PMC article.
-
Clinical impact of bronchoalveolar lavage fluid metagenomic next-generation sequencing in immunocompromised patients with severe community-acquired pneumonia in ICU: a multicenter retrospective study.Infection. 2025 Apr 23. doi: 10.1007/s15010-025-02520-0. Online ahead of print. Infection. 2025. PMID: 40268850
-
Identification of Pathogens in HIV-Infected Patients Using Metagenomic Next-Generation Sequencing (mNGS) as Compared to Conventional Microbiological Tests (CMTs).Infect Drug Resist. 2025 Feb 17;18:929-940. doi: 10.2147/IDR.S491946. eCollection 2025. Infect Drug Resist. 2025. PMID: 39990777 Free PMC article.
-
Application of Targeted Next-Generation Sequencing in Bronchoalveolar Lavage Fluid for the Detection of Pathogens in Pulmonary Infections.Infect Drug Resist. 2025 Jan 27;18:511-522. doi: 10.2147/IDR.S499265. eCollection 2025. Infect Drug Resist. 2025. PMID: 39898354 Free PMC article.
References
-
- Khan MN. Global, regional, and national incidence and mortality burden of non-COVID-19 lower respiratory infections and aetiologies, 1990–2021: a systematic analysis from the global burden of Disease Study 2021 GBD 2021. 2024. - PubMed
-
- José RJ. Respiratory infections: a global burden. Annals Res Hosp 2018, 2.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

