PNSC928, a plant-derived compound, specifically disrupts CtBP2-p300 interaction and reduces inflammation in mice with acute respiratory distress syndrome
- PMID: 38902802
- PMCID: PMC11191317
- DOI: 10.1186/s13062-024-00491-0
PNSC928, a plant-derived compound, specifically disrupts CtBP2-p300 interaction and reduces inflammation in mice with acute respiratory distress syndrome
Abstract
Background: Prior research has highlighted the involvement of a transcriptional complex comprising C-terminal binding protein 2 (CtBP2), histone acetyltransferase p300, and nuclear factor kappa B (NF-κB) in the transactivation of proinflammatory cytokine genes, contributing to inflammation in mice with acute respiratory distress syndrome (ARDS). Nonetheless, it remains uncertain whether the therapeutic targeting of the CtBP2-p300-NF-κB complex holds potential for ARDS suppression.
Methods: An ARDS mouse model was established using lipopolysaccharide (LPS) exposure. RNA-Sequencing (RNA-Seq) was performed on ARDS mice and LPS-treated cells with CtBP2, p300, and p65 knockdown. Small molecules inhibiting the CtBP2-p300 interaction were identified through AlphaScreen. Gene and protein expression levels were quantified using RT-qPCR and immunoblots. Tissue damage was assessed via histological staining.
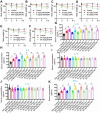
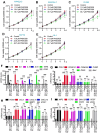
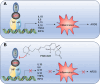
Key findings: We elucidated the specific role of the CtBP2-p300-NF-κB complex in proinflammatory gene regulation. RNA-seq analysis in LPS-challenged ARDS mice and LPS-treated CtBP2-knockdown (CtBP2KD), p300KD, and p65KD cells revealed its significant impact on proinflammatory genes with minimal effects on other NF-κB targets. Commercial inhibitors for CtBP2, p300, or NF-κB exhibited moderate cytotoxicity in vitro and in vivo, affecting both proinflammatory genes and other targets. We identified a potent inhibitor, PNSC928, for the CtBP2-p300 interaction using AlphaScreen. PNSC928 treatment hindered the assembly of the CtBP2-p300-NF-κB complex, substantially downregulating proinflammatory cytokine gene expression without observable cytotoxicity in normal cells. In vivo administration of PNSC928 significantly reduced CtBP2-driven proinflammatory gene expression in ARDS mice, alleviating inflammation and lung injury, ultimately improving ARDS prognosis.
Conclusion: Our results position PNSC928 as a promising therapeutic candidate to specifically target the CtBP2-p300 interaction and mitigate inflammation in ARDS management.
Keywords: ARDS; CtBP2; PNSC928; Proinflammatory cytokine genes; p300.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

