This is a preprint.
Single-nucleus RNA/ATAC-seq in early-stage HCM models predicts SWI/SNF-activation in mutant-myocytes, and allele-specific differences in fibroblasts
- PMID: 38903075
- PMCID: PMC11188105
- DOI: 10.1101/2024.04.24.589078
Single-nucleus RNA/ATAC-seq in early-stage HCM models predicts SWI/SNF-activation in mutant-myocytes, and allele-specific differences in fibroblasts
Abstract
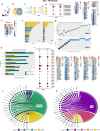
Hypertrophic cardiomyopathy (HCM) is associated with phenotypic variability. To gain insights into transcriptional regulation of cardiac phenotype, single-nucleus linked RNA-/ATAC-seq was performed in 5-week-old control mouse-hearts (WT) and two HCM-models (R92W-TnT, R403Q-MyHC) that exhibit differences in heart size/function and fibrosis; mutant data was compared to WT. Analysis of 23,304 nuclei from mutant hearts, and 17,669 nuclei from WT, revealed similar dysregulation of gene expression, activation of AP-1 TFs (FOS, JUN) and the SWI/SNF complex in both mutant ventricular-myocytes. In contrast, marked differences were observed between mutants, for gene expression/TF enrichment, in fibroblasts, macrophages, endothelial cells. Cellchat predicted activation of pro-hypertrophic IGF-signaling in both mutant ventricular-myocytes, and profibrotic TGFβ-signaling only in mutant-TnT fibroblasts. In summary, our bioinformatics analyses suggest that activation of IGF-signaling, AP-1 TFs and the SWI/SNF chromatin remodeler complex promotes myocyte hypertrophy in early-stage HCM. Selective activation of TGFβ-signaling in mutant-TnT fibroblasts contributes to genotype-specific differences in cardiac fibrosis.
Conflict of interest statement
Competing Interests statement The authors declare no competing interests.
Figures






Similar articles
-
Differences in microRNA-29 and Pro-fibrotic Gene Expression in Mouse and Human Hypertrophic Cardiomyopathy.Front Cardiovasc Med. 2019 Dec 17;6:170. doi: 10.3389/fcvm.2019.00170. eCollection 2019. Front Cardiovasc Med. 2019. PMID: 31921893 Free PMC article.
-
Allele-specific dysregulation of lipid and energy metabolism in early-stage hypertrophic cardiomyopathy.J Mol Cell Cardiol Plus. 2024 Jun;8:100073. doi: 10.1016/j.jmccpl.2024.100073. Epub 2024 Mar 31. J Mol Cell Cardiol Plus. 2024. PMID: 39430912 Free PMC article.
-
Differences in molecular phenotype in mouse and human hypertrophic cardiomyopathy.Sci Rep. 2021 Jun 23;11(1):13163. doi: 10.1038/s41598-021-89451-6. Sci Rep. 2021. PMID: 34162896 Free PMC article.
-
Emerging Themes in Mechanisms of Tumorigenesis by SWI/SNF Subunit Mutation.Epigenet Insights. 2022 Jul 26;15:25168657221115656. doi: 10.1177/25168657221115656. eCollection 2022. Epigenet Insights. 2022. PMID: 35911061 Free PMC article. Review.
-
Familial hypertrophic cardiomyopathy: a paradigm of the cardiac hypertrophic response to injury.Ann Med. 1998 Aug;30 Suppl 1:24-32. Ann Med. 1998. PMID: 9800880 Review.
References
-
- Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–20. - PubMed
-
- Maron BJ, Wolfson JK, Epstein SE and Roberts WC. Intramural ("small vessel") coronary artery disease in hypertrophic cardiomyopathy. J Am Coll Cardiol. 1986;8:545–57. - PubMed
-
- Yalcin H, Valenta I, Yalcin F, Corona-Villalobos C, Vasquez N, Ra J, Kucukler N, Tahari A, Pozios I, Zhou Y, Pomper M, Abraham TP, Schindler TH and Abraham MR. Effect of Diffuse Subendocardial Hypoperfusion on Left Ventricular Cavity Size by (13)N-Ammonia Perfusion PET in Patients With Hypertrophic Cardiomyopathy. Am J Cardiol. 2016;118:1908–1915. - PubMed
-
- Sivalokanathan S, Zghaib T, Greenland GV, Vasquez N, Kudchadkar SM, Kontari E, Lu DY, Dolores-Cerna K, van der Geest RJ, Kamel IR, Olgin JE, Abraham TP, Nazarian S, Zimmerman SL and Abraham MR. Hypertrophic Cardiomyopathy Patients With Paroxysmal Atrial Fibrillation Have a High Burden of Left Atrial Fibrosis by Cardiac Magnetic Resonance Imaging. JACC Clin Electrophysiol. 2019;5:364–375. - PubMed
-
- Lu DY, Ventoulis I, Liu H, Kudchadkar SM, Greenland GV, Yalcin H, Kontari E, Goyal S, Corona-Villalobos CP, Vakrou S, Zimmerman SL, Abraham TP and Abraham MR. Sex-specific cardiac phenotype and clinical outcomes in patients with hypertrophic cardiomyopathy. Am Heart J. 2020;219:58–69. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
