This is a preprint.
Engineering Tumor Stroma Morphogenesis Using Dynamic Cell-Matrix Spheroid Assembly
- PMID: 38903106
- PMCID: PMC11188064
- DOI: 10.1101/2024.03.19.585805
Engineering Tumor Stroma Morphogenesis Using Dynamic Cell-Matrix Spheroid Assembly
Update in
-
A 3D Self-Assembly Platform Integrating Decellularized Matrix Recapitulates In Vivo Tumor Phenotypes and Heterogeneity.Cancer Res. 2025 May 2;85(9):1577-1595. doi: 10.1158/0008-5472.CAN-24-1954. Cancer Res. 2025. PMID: 39888317
Abstract
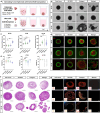
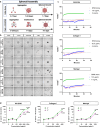
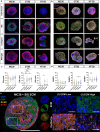
The tumor microenvironment consists of resident tumor cells organized within a compositionally diverse, three-dimensional (3D) extracellular matrix (ECM) network that cannot be replicated in vitro using bottom-up synthesis. We report a new self-assembly system to engineer ECM-rich 3D MatriSpheres wherein tumor cells actively organize and concentrate microgram quantities of decellularized ECM dispersions which modulate cell phenotype. 3D colorectal cancer (CRC) MatriSpheres were created using decellularized small intestine submucosa (SIS) as an orthotopic ECM source that had greater proteomic homology to CRC tumor ECM than traditional ECM formulations such as Matrigel. SIS ECM was rapidly concentrated from its environment and assembled into ECM-rich 3D stroma-like regions by mouse and human CRC cell lines within 4-5 days via a mechanism that was rheologically distinct from bulk hydrogel formation. Both ECM organization and transcriptional regulation by 3D ECM cues affected programs of malignancy, lipid metabolism, and immunoregulation that corresponded with an in vivo MC38 tumor cell subpopulation identified via single cell RNA sequencing. This 3D modeling approach stimulates tumor specific tissue morphogenesis that incorporates the complexities of both cancer cell and ECM compartments in a scalable, spontaneous assembly process that may further facilitate precision medicine.
Keywords: 3D in vitro modeling; biomaterials; decellularization; extracellular matrix; spheroids; tumor microenvironment.
Conflict of interest statement
Competing interests: None
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
