Proliferating macrophages in human tumours show characteristics of monocytes responding to myelopoietic growth factors
- PMID: 38903497
- PMCID: PMC11188303
- DOI: 10.3389/fimmu.2024.1412076
Proliferating macrophages in human tumours show characteristics of monocytes responding to myelopoietic growth factors
Abstract
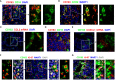
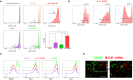
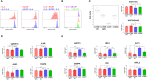
Macrophages play essential roles in maintaining tissue homeostasis and immune defence. However, their extensive infiltration into tumours has been linked to adverse outcomes in multiple human cancers. Within the tumour microenvironment (TME), tumour-associated macrophages (TAMs) promote tumour growth and metastasis, making them prime targets for cancer immunotherapy. Recent single-cell analysis suggest that proliferating TAMs accumulate in human cancers, yet their origins and differentiation pathways remain uncertain. Here, we show that a subpopulation of CD163+ TAMs proliferates in situ within the TME of melanoma, lung cancer, and breast cancer. Consistent with their potential role in suppressing anti-tumour activities of T cells, CD163+ TAMs express a range of potent immunosuppressive molecules, including PD-L1, PD-L2, IL-10, and TGF-β. Other phenotypic markers strongly suggested that these cells originate from CD14+ CCR2+ monocytes, a cell population believed to have minimal capacity for proliferation. However, we demonstrate in vitro that certain myelopoietic cytokines commonly available within the TME induce robust proliferation of human monocytes, especially the combination of interleukin 3 (IL-3) and Macrophage Colony-Stimulating Factor 1 (M-CSF). Monocytic cells cultured with these cytokines efficiently modulate T cell proliferation, and their molecular phenotype recapitulates that of CD163+ TAMs. IL-3-driven proliferation of monocytic cells can be completely blocked by IL-4, associated with the induction of CDKN1A, alongside the upregulation of transcription factors linked to dendritic cell function, such as BATF3 and IRF4. Taken together, our work suggests several novel therapeutic routes to reducing immunosuppressive TAMs in human tumours, from blocking chemokine-mediated recruitment of monocytes to blocking their proliferation.
Keywords: immunosuppression; macrophage proliferation; melanoma; tumour microenvironment; tumour-associated macrophages.
Copyright © 2024 Park, Chen, Verdon, Ooi, Brooks, Martin, Mathy, Emanuel and Dunbar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

