SETD7 Promotes Cell Proliferation and Migration via Methylation-mediated TAF7 in Clear Cell Renal Cell Carcinoma
- PMID: 38904013
- PMCID: PMC11186372
- DOI: 10.7150/ijbs.93201
SETD7 Promotes Cell Proliferation and Migration via Methylation-mediated TAF7 in Clear Cell Renal Cell Carcinoma
Abstract
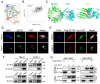
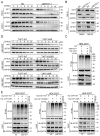
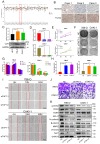
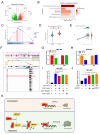
SET domain containing 7(SETD7), a member of histone methyltransferases, is abnormally expressed in multiple tumor types. However, the biological function and underlying molecular mechanism of SETD7 in clear cell renal cell carcinoma (ccRCC) remain unclear. Here, we explored the biological effects of SETD7-TAF7-CCNA2 axis on proliferation and metastasis in ccRCC. We identified both SETD7 and TAF7 were up-regulated and significantly promoted the proliferation and migration of ccRCC cells. Concurrently, there was a significant positive correlation between the expression of SETD7 and TAF7, and the two were colocalized in the nucleus. Mechanistically, SETD7 methylates TAF7 at K5 and K300 sites, resulting in the deubiquitination and stabilization of TAF7. Furthermore, re-expression of TAF7 could partially restore SETD7 knockdown inhibited ccRCC cells proliferation and migration. In addition, TAF7 transcriptionally activated to drive the expression of cyclin A2 (CCNA2). And more importantly, the methylation of TAF7 at K5 and K300 sites exhibited higher transcriptional activity of CCNA2, which promotes formation and progression of ccRCC. Our findings reveal a unique mechanism that SETD7 mediated TAF7 methylation in regulating transcriptional activation of CCNA2 in ccRCC progression and provide a basis for developing effective therapeutic strategies by targeting members of SETD7-TAF7-CCNA2 axis.
Keywords: SETD7; TAF7; clear cell renal cell carcinoma; lysine methylation; oncogene.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




Similar articles
-
NOP2-mediated 5-methylcytosine modification of APOL1 messenger RNA activates PI3K-Akt and facilitates clear cell renal cell carcinoma progression.Int J Biol Sci. 2024 Sep 9;20(12):4853-4871. doi: 10.7150/ijbs.97503. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309431 Free PMC article.
-
NSD1 Inactivation and SETD2 Mutation Drive a Convergence toward Loss of Function of H3K36 Writers in Clear Cell Renal Cell Carcinomas.Cancer Res. 2017 Sep 15;77(18):4835-4845. doi: 10.1158/0008-5472.CAN-17-0143. Epub 2017 Jul 28. Cancer Res. 2017. PMID: 28754676 Free PMC article.
-
CBX4 transcriptionally suppresses KLF6 via interaction with HDAC1 to exert oncogenic activities in clear cell renal cell carcinoma.EBioMedicine. 2020 Mar;53:102692. doi: 10.1016/j.ebiom.2020.102692. Epub 2020 Feb 26. EBioMedicine. 2020. PMID: 32113161 Free PMC article.
-
Lysine N-methyltransferase SETD7 promotes bladder cancer progression and immune escape via STAT3/PD-L1 cascade.Int J Biol Sci. 2023 Jul 16;19(12):3744-3761. doi: 10.7150/ijbs.87182. eCollection 2023. Int J Biol Sci. 2023. PMID: 37564199 Free PMC article.
-
Lysine methyltransferase SETD7 in cancer: functions, molecular mechanisms and therapeutic implications.Mol Biol Rep. 2025 Apr 15;52(1):389. doi: 10.1007/s11033-025-10494-3. Mol Biol Rep. 2025. PMID: 40232640 Review.
Cited by
-
Busulfan Chemotherapy Downregulates TAF7/TNF-α Signaling in Male Germ Cell Dysfunction.Biomedicines. 2024 Sep 28;12(10):2220. doi: 10.3390/biomedicines12102220. Biomedicines. 2024. PMID: 39457533 Free PMC article.
References
-
- Escudier B, Porta C, Schmidinger M, Rioux-Leclercq N, Bex A, Khoo V. et al. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann Oncol. 2019;30:706–20. - PubMed
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. - PubMed
-
- National Cancer C, Renal Cancer Expert Committee of National Cancer Quality Control C. [Quality control index for standardized diagnosis and treatment of renal cancer in China (2022 edition)] Zhonghua Zhong Liu Za Zhi. 2022;44:1256–61. - PubMed
-
- Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z. 2004 WHO classification of the renal tumors of the adults. Eur Urol. 2006;49:798–805. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

