Novel requirements for HAP2/GCS1-mediated gamete fusion in Tetrahymena
- PMID: 38904066
- PMCID: PMC11187246
- DOI: 10.1016/j.isci.2024.110146
Novel requirements for HAP2/GCS1-mediated gamete fusion in Tetrahymena
Abstract
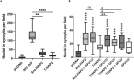
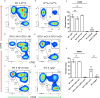
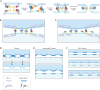
The ancestral gamete fusion protein, HAP2/GCS1, plays an essential role in fertilization in a broad range of taxa. To identify factors that may regulate HAP2/GCS1 activity, we screened mutants of the ciliate Tetrahymena thermophila for behaviors that mimic Δhap2/gcs1 knockout phenotypes in this species. Using this approach, we identified two new genes, GFU1 and GFU2, whose products are necessary for membrane pore formation following mating type recognition and adherence. GFU2 is predicted to be a single-pass transmembrane protein, while GFU1, though lacking obvious transmembrane domains, has the potential to interact directly with membrane phospholipids in the cytoplasm. Like Tetrahymena HAP2/GCS1, expression of GFU1 is required in both cells of a mating pair for efficient fusion to occur. To explain these bilateral requirements, we propose a model that invokes cooperativity between the fusion machinery on apposed membranes of mating cells and accounts for successful fertilization in Tetrahymena's multiple mating type system.
Keywords: Cell biology; Molecular biology.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures

Update of
-
Novel requirements for HAP2-mediated gamete fusion in Tetrahymena.Res Sq [Preprint]. 2023 May 15:rs.3.rs-2928984. doi: 10.21203/rs.3.rs-2928984/v1. Res Sq. 2023. Update in: iScience. 2024 May 28;27(6):110146. doi: 10.1016/j.isci.2024.110146. PMID: 37292678 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

