Dynamic antimicrobial resistance and phylogenomic structure of Salmonella Typhimurium from 2007 to 2019 in Shanghai, China
- PMID: 38904374
- PMCID: PMC11302141
- DOI: 10.1128/spectrum.00262-24
Dynamic antimicrobial resistance and phylogenomic structure of Salmonella Typhimurium from 2007 to 2019 in Shanghai, China
Abstract
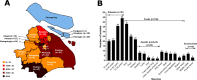
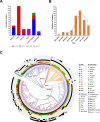
Salmonella enterica serovar Typhimurium is an important foodborne pathogen associated with human salmonellosis worldwide. A retrospective screening was performed to elucidate the prevalence, antimicrobial resistance, and phylogenomic characterization of this pathogen in Shanghai, China. S. Typhimurium isolates were selected from 2,211 serotyped Salmonella isolates collected during 2007-2019. Two hundred and seventy-seven S. Typhimurium isolates were detected in 15 of 16 districts in Shanghai. It was noted that 214 (77.3%) isolates were multi-drug resistant and 32 (11.6%) isolates were resistant to ciprofloxacin and 5 (1.8%) isolates were further resistant to ceftriaxone. Poisson generalized linear mixed model results showed that the multi-drug resistance (MDR) in 2017 and 2018 was significantly higher than that in 2010 (P<0.05), highlighting an increase in the risk of MDR. Phylogenetic results showed that a global data set of 401 sequenced S. Typhimurium isolates was classified into four clones (ST36, ST313, ST19, and ST34), which appeared in international clonal dissemination. The ST34 isolates from China fell into two clades, ST34C1 and ST34C2, the latter of which might originate from Shanghai, and then expanded nationally, accompanied by extended-spectrum β-lactamase gene blaCTX-M-14 and a mutation in quinolone resistance-determining region of the gyrA 87 site. Furthermore, blaCTX-M-14 linking to ISEcp1 upstream and ΔIS903B downstream was found in IncI (Gamma)-like plasmids, and the plasmid conjugation contributed to its horizontal transmission. To our knowledge, it is the first report of the epidemiological and phylogenetic characterization for S. Typhimurium including the emerged clade ST34C2 in Shanghai, warranting the necessity of surveillance for this high-risk pathogen.
Importance: Our study uncovered a widespread distribution of Salmonella enterica serovar Typhimurium isolates in Shanghai accompanied by the increase in antimicrobial resistance (AMR) especially MDR during a 10-year period, which filled in the gap about a long period of continuous monitoring of AMR in this pathogen in Shanghai. Meanwhile, we identified a new clade ST34C2 of S. Typhimurium with the acquisition of IncI (Gamma)-like plasmids mediated by extended-spectrum β-lactamase gene blaCTX-M-14 as well as gyrA 87 mutation, which had not been reported before. It was noted that IncI (Gamma)-like plasmids were reported in S. Typhimurium for the first time and conjugation could accelerate the spread of antimicrobial resistance gene blaCTX-M-14. These findings on the epidemic, antimicrobial resistance, and phylogenomic characterization for S. Typhimurium provide valuable insights into its potential risk to public health and also the basis for AMR prevention and control strategies in Shanghai in the future.
Keywords: IncI (Gamma) plasmids; Salmonella Typhimurium; antimicrobial resistance; phylogenomic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Liang Z, Ke B, Deng X, Liang J, Ran L, Lu L, He D, Huang Q, Ke C, Li Z, Yu H, Klena JD, Wu S. 2015. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong province,China, 2009-2012.. BMC Infect Dis 15. doi:10.1186/s12879-015-0784-4 - DOI - PMC - PubMed
-
- Liang B, Xie Y, He S, Mai J, Huang Y, Yang L, Zhong H, Deng Q, Yao S, Long Y, Yang Y, Gong S, Zhou Z. 2019. Prevalence, serotypes, and drug resistance of nontyphoidal Salmonella among paediatric patients in a tertiary hospital in Guangzhou,China, 2014-2016. J Infect Public Health 12:252–257. doi:10.1016/j.jiph.2018.10.012 - DOI - PubMed
-
- Greening BJ, Whitham HK, Aldous WK, Hall N, Garvey A, Mandernach S, Kahn EB, Nonnenmacher P, Snow J, Meltzer MI, Hoffmann S. 2022. Public health response to multistate Salmonella Typhimurium outbreak associated with prepackaged chicken salad,United States, 2018. Emerg Infect Dis 28:1254–1256. doi:10.3201/eid2806.211633 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

