Neurobeachin regulates hematopoietic progenitor differentiation and survival by modulating Notch activity
- PMID: 38905595
- PMCID: PMC11345395
- DOI: 10.1182/bloodadvances.2023012426
Neurobeachin regulates hematopoietic progenitor differentiation and survival by modulating Notch activity
Abstract
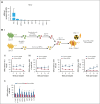
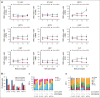
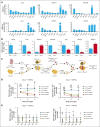
Hematopoietic stem cells (HSCs) can generate all blood cells. This ability is exploited in HSC transplantation (HSCT) to treat hematologic disease. A clear understanding of the molecular mechanisms that regulate HSCT is necessary to continue improving transplant protocols. We identified the Beige and Chediak-Higashi domain-containing protein (BDCP), Neurobeachin (NBEA), as a putative regulator of HSCT. Here, we demonstrated that NBEA and related BDCPs, including LPS Responsive Beige-Like Anchor Protein (LRBA), Neurobeachin Like 1 (NBEAL1) and Lysosomal Trafficking Regulator (LYST), are required during HSCT to efficiently reconstitute the hematopoietic system of lethally irradiated mice. Nbea knockdown in mouse HSCs induced apoptosis and a differentiation block after transplantation. Nbea deficiency in hematopoietic progenitor cells perturbed the expression of genes implicated in vesicle trafficking and led to changes in NOTCH receptor localization. This resulted in perturbation of the NOTCH transcriptional program, which is required for efficient HSC engraftment. In summary, our findings reveal a novel role for NBEA in the control of NOTCH receptor turnover in hematopoietic cells and supports a model in which BDCP-regulated vesicle trafficking is required for efficient HSCT.
© 2024 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures


References
-
- Purton LE. Adult murine hematopoietic stem cells and progenitors: an update on their identities, functions, and assays. Exp Hematol. 2022;116:1–14. - PubMed
-
- Rydstrom A, Grahn THM, Niroula A, et al. Functional and molecular profiling of hematopoietic stem cells during regeneration. Exp Hematol. 2023;127:40–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

