Preclinical efficacy of the potent, selective menin-KMT2A inhibitor JNJ-75276617 (bleximenib) in KMT2A- and NPM1-altered leukemias
- PMID: 38905635
- PMCID: PMC11419783
- DOI: 10.1182/blood.2023022480
Preclinical efficacy of the potent, selective menin-KMT2A inhibitor JNJ-75276617 (bleximenib) in KMT2A- and NPM1-altered leukemias
Abstract
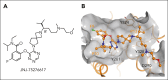
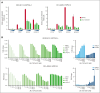
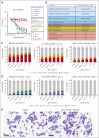
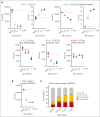
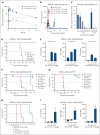
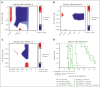
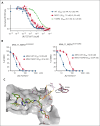
The interaction between menin and histone-lysine N-methyltransferase 2A (KMT2A) is a critical dependency for KMT2A- or nucleophosmin 1 (NPM1)-altered leukemias and an emerging opportunity for therapeutic development. JNJ-75276617 (bleximenib) is a novel, orally bioavailable, potent, and selective protein-protein interaction inhibitor of the binding between menin and KMT2A. In KMT2A-rearranged (KMT2A-r) and NPM1-mutant (NPM1c) acute myeloid leukemia (AML) cells, JNJ-75276617 inhibited the association of the menin-KMT2A complex with chromatin at target gene promoters, resulting in reduced expression of several menin-KMT2A target genes, including MEIS1 and FLT3. JNJ-75276617 displayed potent antiproliferative activity across several AML and acute lymphoblastic leukemia (ALL) cell lines and patient samples harboring KMT2A or NPM1 alterations in vitro. In xenograft models of AML and ALL, JNJ-75276617 reduced leukemic burden and provided a significant dose-dependent survival benefit accompanied by expression changes of menin-KMT2A target genes. JNJ-75276617 demonstrated synergistic effects with gilteritinib in vitro in AML cells harboring KMT2A-r. JNJ-75276617 further exhibited synergistic effects with venetoclax and azacitidine in AML cells bearing KMT2A-r in vitro, and significantly increased survival in mice. Interestingly, JNJ-75276617 showed potent antiproliferative activity in cell lines engineered with recently discovered mutations (MEN1M327I or MEN1T349M) that developed in patients refractory to the menin-KMT2A inhibitor revumenib. A cocrystal structure of menin in complex with JNJ-75276617 indicates a unique binding mode distinct from other menin-KMT2A inhibitors, including revumenib. JNJ-75276617 is being clinically investigated for acute leukemias harboring KMT2A or NPM1 alterations, as a monotherapy for relapsed/refractory acute leukemia (NCT04811560), or in combination with AML-directed therapies (NCT05453903).
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: M.C.K., J.W.T., T.V., V.P., A.M., D.G., P.L.S., F.J., V.K., F.E., B.B., K.V., S.E.A., P.J., L.B., A.K., N.D., D.K., G.U., B.V., G.S.C., R.K., R.S., L.F., C.G., N.D., E.C.P., R.A., Y.E., K.P., and U.P. are currently employees of Janssen Research & Development and may own stock/stock options in Johnson & Johnson. O.Q., X.D., P.V., W.C., J.P.E., and D.M.W. are former employees of Janssen Research & Development and may own stock/stock options in Johnson & Johnson. J.W.T., O.Q., X.D., V.P., W.C., N.D., and G.U. are named as inventors on patent applications related to MENIN inhibition WO/2021/121327. M.C.K., J.W.T., O.Q., X.D., T.V., V.P., W.C., B.B., N.D., L.F., C.G., N.D., E.C.P., K.P., and U.P. are named as inventors on patent applications related to MENIN inhibition WO/2022/237719 and WO/2022/237720. S.A.A. has been a consultant and/or shareholder for Neomorph Inc, Imago Biosciences, Cyteir Therapeutics, C4 Therapeutics, Nimbus Therapeutics, and Accent Therapeutics. S.A.A. has received research support from Janssen and Syndax. S.A.A. is named as an inventor on a patent application related to MENIN inhibition WO/2017/132398A1. E.S.F. is a founder, scientific advisory board member, and equity holder of Civetta Therapeutics, Proximity Therapeutics, and Neomorph, Inc (also board of directors). He is an equity holder and scientific advisory board member for Avilar Therapeutics, Photys Therapeutics, and Ajax Therapeutics and an equity holder in Lighthorse Therapeutics. E.S.F. is a consultant to Novartis, EcoR1 capital, Odyssey, and Deerfield. The Fischer laboratory receives or has received research funding from Deerfield, Novartis, Ajax, Interline, Bayer, and Astellas. The remaining authors declare no competing financial interests.
Figures


Comment in
-
A menin-KMT2A inhibitor to overcome resistance.Blood. 2024 Sep 12;144(11):1139-1140. doi: 10.1182/blood.2024025760. Blood. 2024. PMID: 39264614 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

