Adult microglial TGFβ1 is required for microglia homeostasis via an autocrine mechanism to maintain cognitive function in mice
- PMID: 38906887
- PMCID: PMC11192737
- DOI: 10.1038/s41467-024-49596-0
Adult microglial TGFβ1 is required for microglia homeostasis via an autocrine mechanism to maintain cognitive function in mice
Abstract
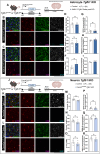
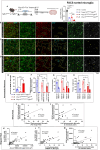
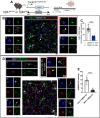
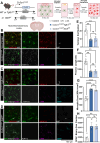
While TGF-β signaling is essential for microglial function, the cellular source of TGF-β1 ligand and its spatial regulation remains unclear in the adult CNS. Our data supports that microglia but not astrocytes or neurons are the primary producers of TGF-β1 ligands needed for microglial homeostasis. Microglia-Tgfb1 KO leads to the activation of microglia featuring a dyshomeostatic transcriptome that resembles disease-associated, injury-associated, and aged microglia, suggesting microglial self-produced TGF-β1 ligands are important in the adult CNS. Astrocytes in MG-Tgfb1 inducible (i)KO mice show a transcriptome profile that is closely aligned with an LPS-associated astrocyte profile. Additionally, using sparse mosaic single-cell microglia KO of TGF-β1 ligand we established an autocrine mechanism for signaling. Here we show that MG-Tgfb1 iKO mice present cognitive deficits, supporting that precise spatial regulation of TGF-β1 ligand derived from microglia is required for the maintenance of brain homeostasis and normal cognitive function in the adult brain.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
Microglia-derived TGF-β1 ligand maintains microglia homeostasis via autocrine mechanism and is critical for normal cognitive function in adult mouse brain.bioRxiv [Preprint]. 2023 Jul 7:2023.07.05.547814. doi: 10.1101/2023.07.05.547814. bioRxiv. 2023. Update in: Nat Commun. 2024 Jun 21;15(1):5306. doi: 10.1038/s41467-024-49596-0. PMID: 37461569 Free PMC article. Updated. Preprint.
References
-
- Carroll JA, Race B, Williams K, Striebel JF, Chesebro B. Innate immune responses after stimulation with toll-like receptor agonists in ex vivo microglial cultures and an in vivo model using mice with reduced microglia. J. Neuroinflammation. 2021;18:194. doi: 10.1186/s12974-021-02240-w. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R35 GM146890/GM/NIGMS NIH HHS/United States
- R21NS127177/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- T32 ES007250/ES/NIEHS NIH HHS/United States
- R01AG083164/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- T32 NS007453/NS/NINDS NIH HHS/United States
- R01 NS125074/NS/NINDS NIH HHS/United States
- F31 NS125930/NS/NINDS NIH HHS/United States
- 1F31NS125930/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R21 NS127177/NS/NINDS NIH HHS/United States
- R01 AG083164/AG/NIA NIH HHS/United States
- R01NS125074/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- 663007/Simons Foundation
- S10 OD030402/OD/NIH HHS/United States
- R35GM146890/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- F31 NS129204/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

