RNA-Seq Data Analysis
- PMID: 38907924
- PMCID: PMC12125953
- DOI: 10.1007/978-1-0716-3918-4_18
RNA-Seq Data Analysis
Abstract
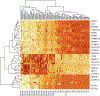
RNA-Seq data analysis stands as a vital part of genomics research, turning vast and complex datasets into meaningful biological insights. It is a field marked by rapid evolution and ongoing innovation, necessitating a thorough understanding for anyone seeking to unlock the potential of RNA-Seq data. In this chapter, we describe the intricate landscape of RNA-seq data analysis, elucidating a comprehensive pipeline that navigates through the entirety of this complex process. Beginning with quality control, the chapter underscores the paramount importance of ensuring the integrity of RNA-seq data, as it lays the groundwork for subsequent analyses. Preprocessing is then addressed, where the raw sequence data undergoes necessary modifications and enhancements, setting the stage for the alignment phase. This phase involves mapping the processed sequences to a reference genome, a step pivotal for decoding the origins and functions of these sequences.Venturing into the heart of RNA-seq analysis, the chapter then explores differential expression analysis-the process of identifying genes that exhibit varying expression levels across different conditions or sample groups. Recognizing the biological context of these differentially expressed genes is pivotal; hence, the chapter transitions into functional analysis. Here, methods and tools like Gene Ontology and pathway analyses help contextualize the roles and interactions of the identified genes within broader biological frameworks. However, the chapter does not stop at conventional analysis methods. Embracing the evolving paradigms of data science, it delves into machine learning applications for RNA-seq data, introducing advanced techniques in dimension reduction and both unsupervised and supervised learning. These approaches allow for patterns and relationships to be discerned in the data that might be imperceptible through traditional methods.
Keywords: Differential expression; Next-generation sequencing; Sequence alignment; Transcriptomics.
© 2024. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Figures
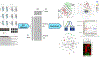

References
-
- Garber M, Grabherr MG, Guttman M, et al. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nature methods, 8(6): 469–477. - PubMed
-
- Martin JA, Wang Z, (2011). Next-generation transcriptome assembly. Nature Reviews Genetics, 12(10): 671–682. - PubMed
-
- Wang L, Wang S, Li W, (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics, 28(16): 2184–2185. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

