DiPPI: A Curated Data Set for Drug-like Molecules in Protein-Protein Interfaces
- PMID: 38907989
- PMCID: PMC11577314
- DOI: 10.1021/acs.jcim.3c01905
DiPPI: A Curated Data Set for Drug-like Molecules in Protein-Protein Interfaces
Abstract
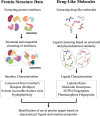
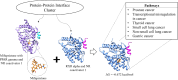
Proteins interact through their interfaces, and dysfunction of protein-protein interactions (PPIs) has been associated with various diseases. Therefore, investigating the properties of the drug-modulated PPIs and interface-targeting drugs is critical. Here, we present a curated large data set for drug-like molecules in protein interfaces. We further introduce DiPPI (Drugs in Protein-Protein Interfaces), a two-module web site to facilitate the search for such molecules and their properties by exploiting our data set in drug repurposing studies. In the interface module of the web site, we present several properties, of interfaces, such as amino acid properties, hotspots, evolutionary conservation of drug-binding amino acids, and post-translational modifications of these residues. On the drug-like molecule side, we list drug-like small molecules and FDA-approved drugs from various databases and highlight those that bind to the interfaces. We further clustered the drugs based on their molecular fingerprints to confine the search for an alternative drug to a smaller space. Drug properties, including Lipinski's rules and various molecular descriptors, are also calculated and made available on the web site to guide the selection of drug molecules. Our data set contains 534,203 interfaces for 98,632 protein structures, of which 55,135 are detected to bind to a drug-like molecule. 2214 drug-like molecules are deposited on our web site, among which 335 are FDA-approved. DiPPI provides users with an easy-to-follow scheme for drug repurposing studies through its well-curated and clustered interface and drug data and is freely available at http://interactome.ku.edu.tr:8501.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces.BMC Bioinformatics. 2006 Mar 2;7:104. doi: 10.1186/1471-2105-7-104. BMC Bioinformatics. 2006. PMID: 16512892 Free PMC article.
-
HotSprint: database of computational hot spots in protein interfaces.Nucleic Acids Res. 2008 Jan;36(Database issue):D662-6. doi: 10.1093/nar/gkm813. Epub 2007 Oct 24. Nucleic Acids Res. 2008. PMID: 17959648 Free PMC article.
-
Predicting where small molecules bind at protein-protein interfaces.PLoS One. 2013;8(3):e58583. doi: 10.1371/journal.pone.0058583. Epub 2013 Mar 7. PLoS One. 2013. PMID: 23505538 Free PMC article.
-
In silico structure-based approaches to discover protein-protein interaction-targeting drugs.Methods. 2017 Dec 1;131:22-32. doi: 10.1016/j.ymeth.2017.08.006. Epub 2017 Aug 9. Methods. 2017. PMID: 28802714 Free PMC article. Review.
-
Drugs targeting protein-protein interactions.ChemMedChem. 2006 Apr;1(4):400-11. doi: 10.1002/cmdc.200600004. ChemMedChem. 2006. PMID: 16892375 Review.
References
-
- Pushpakom S.; Iorio F.; Eyers P. A.; Escott K. J.; Hopper S.; Wells A.; Doig A.; Guilliams T.; Latimer J.; McNamee C.; Norris A.; Sanseau P.; Cavalla D.; Pirmohamed M. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discovery 2019, 18 (1), 41–58. 10.1038/nrd.2018.168. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

