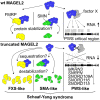
Truncated variants of MAGEL2 are involved in the etiologies of the Schaaf-Yang and Prader-Willi syndromes
- PMID: 38908375
- PMCID: PMC11267527
- DOI: 10.1016/j.ajhg.2024.05.023
Truncated variants of MAGEL2 are involved in the etiologies of the Schaaf-Yang and Prader-Willi syndromes
Erratum in
-
Truncated variants of MAGEL2 are involved in the etiologies of the Schaaf-Yang and Prader-Willi syndromes.Am J Hum Genet. 2025 Oct 2;112(10):2562. doi: 10.1016/j.ajhg.2025.09.005. Epub 2025 Sep 12. Am J Hum Genet. 2025. PMID: 40945516 No abstract available.
Abstract
The neurodevelopmental disorders Prader-Willi syndrome (PWS) and Schaaf-Yang syndrome (SYS) both arise from genomic alterations within human chromosome 15q11-q13. A deletion of the SNORD116 cluster, encoding small nucleolar RNAs, or frameshift mutations within MAGEL2 result in closely related phenotypes in individuals with PWS or SYS, respectively. By investigation of their subcellular localization, we observed that in contrast to a predominant cytoplasmic localization of wild-type (WT) MAGEL2, a truncated MAGEL2 mutant was evenly distributed between the cytoplasm and the nucleus. To elucidate regulatory pathways that may underlie both diseases, we identified protein interaction partners for WT or mutant MAGEL2, in particular the survival motor neuron protein (SMN), involved in spinal muscular atrophy, and the fragile-X-messenger ribonucleoprotein (FMRP), involved in autism spectrum disorders. The interactome of the non-coding RNA SNORD116 was also investigated by RNA-CoIP. We show that WT and truncated MAGEL2 were both involved in RNA metabolism, while regulation of transcription was mainly observed for WT MAGEL2. Hence, we investigated the influence of MAGEL2 mutations on the expression of genes from the PWS locus, including the SNORD116 cluster. Thereby, we provide evidence for MAGEL2 mutants decreasing the expression of SNORD116, SNORD115, and SNORD109A, as well as protein-coding genes MKRN3 and SNRPN, thus bridging the gap between PWS and SYS.
Keywords: Schaaf-Yang syndrome, Prader-Willi syndrome, MAGEL2, SNORD116, SMN, FMRP.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Szabadi S., Sila Z., Dewey J., Rowland D., Penugonda M., Ergun-Longmire B. A Review of Prader–Willi Syndrome. Endocrines. 2022;3:329–348. doi: 10.3390/endocrines3020027. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

