Development of a single-chain variable antibody fragment against a conserved region of the SARS-CoV-2 spike protein
- PMID: 38909102
- PMCID: PMC11193732
- DOI: 10.1038/s41598-024-64103-7
Development of a single-chain variable antibody fragment against a conserved region of the SARS-CoV-2 spike protein
Abstract
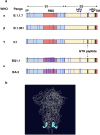
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has prolonged the duration of the pandemic because of the continuous emergence of new variant strains. The emergence of these mutant strains makes it difficult to detect the virus with the existing antibodies; thus, the development of novel antibodies that can target both the variants as well as the original strain is necessary. In this study, we generated a high-affinity monoclonal antibody (5G2) against the highly conserved region of the SARS-CoV-2 spike protein to detect the protein variants. Moreover, we generated its single-chain variable antibody fragment (sc5G2). The sc5G2 expressed in mammalian and bacterial cells detected the spike protein of the original SARS-CoV-2 and variant strains. The resulting sc5G2 will be a useful tool to detect the original SARS-CoV-2 and variant strains.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Generation and utility of a single-chain fragment variable monoclonal antibody platform against a baculovirus expressed recombinant receptor binding domain of SARS-CoV-2 spike protein.Mol Immunol. 2022 Jan;141:287-296. doi: 10.1016/j.molimm.2021.12.006. Epub 2021 Dec 10. Mol Immunol. 2022. PMID: 34915268 Free PMC article.
-
Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.MAbs. 2021 Jan-Dec;13(1):1953683. doi: 10.1080/19420862.2021.1953683. MAbs. 2021. PMID: 34313527 Free PMC article.
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
Research progress of spike protein mutation of SARS-CoV-2 mutant strain and antibody development.Front Immunol. 2024 Nov 18;15:1407149. doi: 10.3389/fimmu.2024.1407149. eCollection 2024. Front Immunol. 2024. PMID: 39624100 Free PMC article. Review.
-
An approach towards development of monoclonal IgY antibodies against SARS CoV-2 spike protein (S) using phage display method: A review.Int Immunopharmacol. 2020 Aug;85:106654. doi: 10.1016/j.intimp.2020.106654. Epub 2020 Jun 3. Int Immunopharmacol. 2020. PMID: 32512271 Free PMC article. Review.
References
-
- Fiolet T, Kherabi Y, MacDonald CJ, Ghosn J, Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022;28:202–221. doi: 10.1016/j.cmi.2021.10.005. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

