Integrating metabolomics and transcriptomics to analyze the differences of breast muscle quality and flavor formation between Daweishan mini chicken and broiler
- PMID: 38909504
- PMCID: PMC11253666
- DOI: 10.1016/j.psj.2024.103920
Integrating metabolomics and transcriptomics to analyze the differences of breast muscle quality and flavor formation between Daweishan mini chicken and broiler
Abstract
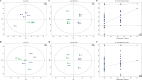
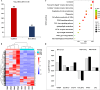
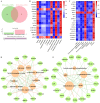
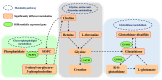
The quality and flavor of chicken are affected by muscle metabolites and related regulatory genes, and the molecular regulation mechanism of meat quality is different among different breeds of chicken. In this study, 40 one-day-old Daweishan mini chicken (DM) and Cobb broiler (CB) were selected from each group, with 4 replicates and 10 chickens in each replicate. The chickens were reared until 90 d of age under the same management conditions. Then, metabolomics and transcriptomics data of 90-day-old DM (n = 4) and CB (n = 4) were integrated to analyze metabolites affecting breast muscle quality and flavor, and to explore the important genes regulating meat quality and flavor related metabolites. The results showed that a total of 38 significantly different metabolites (SDMs) and 420 differentially expressed genes (DEGs) were detected in the breast muscle of the 2 breeds. Amino acid and lipid metabolism may be the cause of meat quality and flavor difference between DM and CB chickens, involving metabolites such as L-methionine, betaine, N6, N6, N6-Trimethyl-L-lysine, L-anserine, glutathione, glutathione disulfide, L-threonine, N-Acetyl-L-aspartic acid, succinate, choline, DOPC, SOPC, alpha-linolenic acid, L-palmitoylcarnitine, etc. Important regulatory genes with high correlation with flavor amino acids (GATM, GSTO1) and lipids (PPARG, LPL, PLIN1, SCD, ANGPTL4, FABP7, GK, B4GALT6, UGT8, PLPP4) were identified by correlation analysis, and the gene-metabolite interaction network of breast muscle mass and flavor formation in DM chicken was constructed. This study showed that there were significant differences in breast metabolites between DM and CB chickens, mainly in amino acid and lipid metabolites. These 2 kinds of substances may be the main reasons for the difference in breast muscle quality and flavor between the 2 breeds. In general, this study could provide a theoretical basis for further research on the molecular regulatory mechanism of the formation of breast muscle quality and flavor differences between DM and CB chickens, and provide a reference for the development, utilization and genetic breeding of high-quality meat chicken breeds.
Keywords: amino acid; flavor compound; lipid; native chicken; regulation mechanism.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
DISCLOSURES The authors declare no conflicts of interest.
Figures

Similar articles
-
Integrating metabolomics and transcriptomics to analyze differences in muscle mass and flavor formation in Gayal and yellow cattle.Front Vet Sci. 2025 May 14;12:1581767. doi: 10.3389/fvets.2025.1581767. eCollection 2025. Front Vet Sci. 2025. PMID: 40438416 Free PMC article.
-
Metabolomic profiles and compositional differences involved in flavor characteristics of raw breast meat from slow- and fast-growing chickens in Thailand.Poult Sci. 2024 Nov;103(11):104230. doi: 10.1016/j.psj.2024.104230. Epub 2024 Aug 23. Poult Sci. 2024. PMID: 39236465 Free PMC article.
-
Integrative metabolomics and transcriptomics analysis revealed specific genes and metabolites affecting meat quality of chickens under different rearing systems.Poult Sci. 2024 Sep;103(9):103994. doi: 10.1016/j.psj.2024.103994. Epub 2024 Jun 24. Poult Sci. 2024. PMID: 38991385 Free PMC article.
-
Unraveling the flavor profiles of chicken meat: Classes, biosynthesis, influencing factors in flavor development, and sensory evaluation.Compr Rev Food Sci Food Saf. 2024 Jul;23(4):e13391. doi: 10.1111/1541-4337.13391. Compr Rev Food Sci Food Saf. 2024. PMID: 39042376 Review.
-
Study of emerging chicken meat quality defects using OMICs: What do we know?J Proteomics. 2023 Mar 30;276:104837. doi: 10.1016/j.jprot.2023.104837. Epub 2023 Feb 11. J Proteomics. 2023. PMID: 36781045 Review.
Cited by
-
Integrating metabolomics and transcriptomics to analyze differences in muscle mass and flavor formation in Gayal and yellow cattle.Front Vet Sci. 2025 May 14;12:1581767. doi: 10.3389/fvets.2025.1581767. eCollection 2025. Front Vet Sci. 2025. PMID: 40438416 Free PMC article.
-
Dual-targeted microbubbles for atherosclerosis therapy: Inducing M1 macrophage apoptosis by inhibiting telomerase activity.Mater Today Bio. 2025 Mar 20;32:101675. doi: 10.1016/j.mtbio.2025.101675. eCollection 2025 Jun. Mater Today Bio. 2025. PMID: 40225135 Free PMC article.
References
-
- Aggrey S.E., González-Cerón F., Rekaya R., Mercier Y. Gene expression differences in the methionine remethylation and transsulphuration pathways under methionine restriction and recovery with D,L-methionine or D,L-HMTBA in meat-type chickens. J. Anim. Physiol. Anim. Nutr (Berl). 2018;102:e468–e475. - PubMed
-
- Alex S., Lange K., Amolo T., Grinstead J.S., Haakonsson A.K., Szalowska E., Koppen A., Mudde K., Haenen D., Al-Lahham S., Roelofsen H., Houtman R., van der Burg B., Mandrup S., Bonvin A.M., Kalkhoven E., Müller M., Hooiveld G.J., Kersten S. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol. Cell. Biol. 2013;33:1303–1316. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

