A small molecule that inhibits the evolution of antibiotic resistance
- PMID: 38911259
- PMCID: PMC11188740
- DOI: 10.1093/narmme/ugae001
A small molecule that inhibits the evolution of antibiotic resistance
Abstract
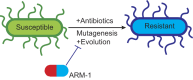
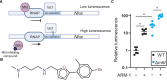
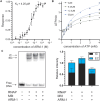
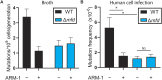
Antibiotic resistance rapidly develops against almost all available therapeutics. Therefore, searching for new antibiotics to overcome the problem of antibiotic resistance alone is insufficient. Given that antibiotic resistance can be driven by mutagenesis, an avenue for preventing it is the inhibition of mutagenic processes. We previously showed that the DNA translocase Mfd is mutagenic and accelerates antibiotic resistance development. Here, we present our discovery of a small molecule that inhibits Mfd-dependent mutagenesis, ARM-1 (anti-resistance molecule 1). We found ARM-1 using a high-throughput, small molecule, in vivo screen. Using biochemical assays, we characterized the mechanism by which ARM-1 inhibits Mfd. Critically, we found that ARM-1 reduces mutagenesis and significantly delays antibiotic resistance development across highly divergent bacterial pathogens. These results demonstrate that the mutagenic proteins accelerating evolution can be directly inhibited. Furthermore, our findings suggest that Mfd inhibition, alongside antibiotics, is a potentially effective approach for prevention of antibiotic resistance development during treatment of infections.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Molecular Medicine.
Figures

References
-
- O’Neill J. The review on antimicrobial resistance: tackling drug-resistant infections globally. Arch. Pharm. Pract. 2016; 7:110.
-
- Beyer P., Paulin S.. The antibacterial research and development pipeline needs urgent solutions. ACS Infect. Dis. 2020; 6:1289–1291.
Grants and funding
LinkOut - more resources
Full Text Sources
