Differences in the Membrane-Binding Properties of Flaviviral Nonstructural 1 (NS1) Protein: Comparative Simulations of Zika and Dengue Virus NS1 Proteins in Explicit Bilayers
- PMID: 38911907
- PMCID: PMC11191575
- DOI: 10.1021/acsbiomedchemau.3c00073
Differences in the Membrane-Binding Properties of Flaviviral Nonstructural 1 (NS1) Protein: Comparative Simulations of Zika and Dengue Virus NS1 Proteins in Explicit Bilayers
Abstract
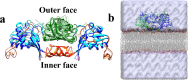
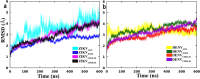
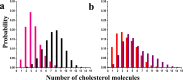
NS1 in flaviviruses is the only nonstructural protein that is secretory and interacts with different cellular components of the host cell membrane. NS1 is localized in the ER as a dimer to facilitate viral replication. Crystal structures of NS1 homologues from zika (ZIKV) and dengue (DENV) viruses have revealed the organization of different domains in NS1 dimers. The β-roll and the connector and intertwined loop regions of wing domains of NS1 have been shown to interact with the membranes. In this study, we have performed multiple molecular dynamics (MD) simulations of ZIKV and DENV NS1 systems in apo and in POPE bilayers with different cholesterol concentrations (0, 20 and 40%). The NS1 protein was placed just above the membrane surface, and for each NS1-membrane system two to three independent simulations with 600 ns production run were performed. At the end of the production runs, ZIKV NS1 inserts deeper inside the membrane compared to the DENV counterpart. Unlike ZIKV NS1, the orientation of DENV NS1 is asymmetric in which one of the chains in the dimer interacts with the membrane while the other is more exposed to the solvent. The β-roll region in ZIKV NS1 penetrates beyond the headgroup region and interacts with the lipid acyl chains while the C-terminal region barely interacts with the headgroup. Specific residues in the intertwined region deeply penetrate inside the membrane. The role of charged and aromatic residues of ZIKV NS1 in strongly interacting with the membrane components is revealed. The presence of cholesterol affects the extent of insertion in the membrane and interaction of individual residues. Overall, membrane-binding properties of ZIKV NS1 significantly differ from its counterpart in DENV. The differences found in the binding and insertion of NS1 can be used to design drugs and novel antibodies that can be flavivirus specific.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Molecular Determinants of Tissue Specificity of Flavivirus Nonstructural Protein 1 Interaction with Endothelial Cells.J Virol. 2022 Oct 12;96(19):e0066122. doi: 10.1128/jvi.00661-22. Epub 2022 Sep 15. J Virol. 2022. PMID: 36106873 Free PMC article.
-
Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure.EMBO J. 2016 Oct 17;35(20):2170-2178. doi: 10.15252/embj.201695290. Epub 2016 Aug 30. EMBO J. 2016. PMID: 27578809 Free PMC article.
-
The Dengue Virus Nonstructural Protein 1 (NS1) Interacts with the Putative Epigenetic Regulator DIDO1 to Promote Flavivirus Replication in Mosquito Cells.J Virol. 2022 Jun 22;96(12):e0070422. doi: 10.1128/jvi.00704-22. Epub 2022 Jun 2. J Virol. 2022. PMID: 35652656 Free PMC article.
-
Zika virus structural biology and progress in vaccine development.Biotechnol Adv. 2018 Jan-Feb;36(1):47-53. doi: 10.1016/j.biotechadv.2017.09.004. Epub 2017 Sep 12. Biotechnol Adv. 2018. PMID: 28916391 Review.
-
Making sense of flavivirus non-strctural protein 1 in innate immune evasion and inducing tissue-specific damage.Virus Res. 2023 Oct 15;336:199222. doi: 10.1016/j.virusres.2023.199222. Epub 2023 Sep 16. Virus Res. 2023. PMID: 37716670 Free PMC article. Review.
Cited by
-
Identification of Immunodominant Epitopes of Dengue Virus 2 Envelope and NS1 Proteins: Evaluating the Diagnostic Potential of a Synthetic Peptide.Mol Diagn Ther. 2024 Sep;28(5):633-643. doi: 10.1007/s40291-024-00728-8. Epub 2024 Jul 9. Mol Diagn Ther. 2024. PMID: 38980575
References
-
- Bhatt S.; Gething P. W.; Brady O. J.; Messina J. P.; Farlow A. W.; Moyes C. L.; Drake J. M.; Brownstein J. S.; Hoen A. G.; Sankoh O.; Myers M. F.; George D. B.; Jaenisch T.; Wint G. R.; Simmons C. P.; Scott T. W.; Farrar J. J.; Hay S. I. The global distribution and burden of dengue. Nature 2013, 496 (7446), 504–7. 10.1038/nature12060. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
