Preanalytical considerations in quantifying circulating miRNAs that predict end-stage kidney disease in diabetes
- PMID: 38912578
- PMCID: PMC11383361
- DOI: 10.1172/jci.insight.174153
Preanalytical considerations in quantifying circulating miRNAs that predict end-stage kidney disease in diabetes
Abstract
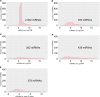
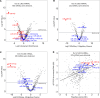
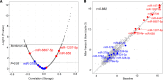
Our previous study identified 8 risk and 9 protective plasma miRNAs associated with progression to end-stage kidney disease (ESKD) in diabetes. This study aimed to elucidate preanalytical factors that influence the quantification of circulating miRNAs. Using the EdgeSeq platform, which quantifies 2,002 miRNAs in plasma, including ESKD-associated miRNAs, we compared miRNA profiles in whole plasma versus miRNA profiles in RNA extracted from the same plasma specimens. Less than half of the miRNAs were detected in standard RNA extraction from plasma. Detection of individual and concentrations of miRNAs were much lower when RNA extracted from plasma was quantified by RNA sequencing (RNA-Seq) or quantitative reverse transcription PCR (qRT-PCR) platforms compared with EdgeSeq. Plasma profiles of miRNAs determined by the EdgeSeq platform had excellent reproducibility in assessment and had no variation with age, sex, hemoglobin A1c, BMI, and cryostorage time. The risk ESKD-associated miRNAs were detected and measured accurately only in whole plasma and using the EdgeSeq platform. Protective ESKD-associated miRNAs were detected by all platforms except qRT-PCR; however, correlations among concentrations obtained with different platforms were weak or nonexistent. In conclusion, preanalytical factors have a profound effect on detection and quantification of circulating miRNAs in ESKD in diabetes. Quantification of miRNAs in whole plasma and using the EdgeSeq platform may be the preferable method to study profiles of circulating cell-free miRNAs associated with ESKD and possibly other diseases.
Keywords: Chronic kidney disease; Diabetes; Nephrology; Noncoding RNAs.
Conflict of interest statement
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

