Growing Glycans in Rosetta: Accurate de novo glycan modeling, density fitting, and rational sequon design
- PMID: 38913746
- PMCID: PMC11288642
- DOI: 10.1371/journal.pcbi.1011895
Growing Glycans in Rosetta: Accurate de novo glycan modeling, density fitting, and rational sequon design
Abstract
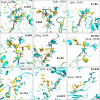
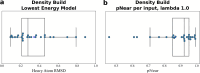
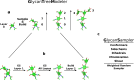
Carbohydrates and glycoproteins modulate key biological functions. However, experimental structure determination of sugar polymers is notoriously difficult. Computational approaches can aid in carbohydrate structure prediction, structure determination, and design. In this work, we developed a glycan-modeling algorithm, GlycanTreeModeler, that computationally builds glycans layer-by-layer, using adaptive kernel density estimates (KDE) of common glycan conformations derived from data in the Protein Data Bank (PDB) and from quantum mechanics (QM) calculations. GlycanTreeModeler was benchmarked on a test set of glycan structures of varying lengths, or "trees". Structures predicted by GlycanTreeModeler agreed with native structures at high accuracy for both de novo modeling and experimental density-guided building. We employed these tools to design de novo glycan trees into a protein nanoparticle vaccine to shield regions of the scaffold from antibody recognition, and experimentally verified shielding. This work will inform glycoprotein model prediction, glycan masking, and further aid computational methods in experimental structure determination and refinement.
Copyright: © 2024 Adolf-Bryfogle et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: Dr. JJG is an unpaid board member of the Rosetta Commons. Under institutional participation agreements between the University of Washington, acting on behalf of the Rosetta Commons, Johns Hopkins University may be entitled to a portion of revenue received on licensing Rosetta software including methods discussed/developed in this study. As a member of the Scientific Advisory Board, JJG has a financial interest in Cyrus Biotechnology. Cyrus Biotechnology distributes the Rosetta software, which may include methods developed in this study. These arrangements have been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. WRS is an employee of Moderna, Inc., but his contributions to this work were all conducted prior to his employment at Moderna. JAB is an employee of Johnson and Johnson Innovative Medicine, Inc., but his contributions to this work were all conducted prior to his current employment.
Figures




References
-
- Ernst B, Hart GW, Sinaý P. Carbohydrates in Chemistry and Biology [Internet]. 1st ed. Wiley; 2000. [cited 2020 Dec 18]. Available from: https://onlinelibrary.wiley.com/doi/book/10.1002/9783527618255. - DOI
-
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, et al.. Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans. Trkola A, editor. PLoS Pathog. 2013. May 2;9(5):e1003342. doi: 10.1371/journal.ppat.1003342 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

