Cancer cell plasticity defines response to immunotherapy in cutaneous squamous cell carcinoma
- PMID: 38914547
- PMCID: PMC11196727
- DOI: 10.1038/s41467-024-49718-8
Cancer cell plasticity defines response to immunotherapy in cutaneous squamous cell carcinoma
Abstract
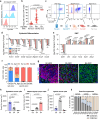
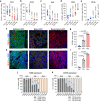
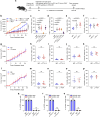
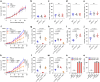
Immune checkpoint blockade (ICB) approaches have changed the therapeutic landscape for many tumor types. However, half of cutaneous squamous cell carcinoma (cSCC) patients remain unresponsive or develop resistance. Here, we show that, during cSCC progression in male mice, cancer cells acquire epithelial/mesenchymal plasticity and change their immune checkpoint (IC) ligand profile according to their features, dictating the IC pathways involved in immune evasion. Epithelial cancer cells, through the PD-1/PD-L1 pathway, and mesenchymal cancer cells, through the CTLA-4/CD80 and TIGIT/CD155 pathways, differentially block antitumor immune responses and determine the response to ICB therapies. Accordingly, the anti-PD-L1/TIGIT combination is the most effective strategy for blocking the growth of cSCCs that contain both epithelial and mesenchymal cancer cells. The expression of E-cadherin/Vimentin/CD80/CD155 proteins in cSCC, HNSCC and melanoma patient samples predicts response to anti-PD-1/PD-L1 therapy. Collectively, our findings indicate that the selection of ICB therapies should take into account the epithelial/mesenchymal features of cancer cells.
© 2024. The Author(s).
Conflict of interest statement
M.E. declares research grants from Ferrer International and Incyte, and consulting fees from Quimatryx outside of this study. M.O. declares consulting or advisory arrangements with Merck, MSD and Transgene; research support (clinical trials) from Merck and Roche; that the institution receives clinical trial support from AbbVie, Ayala Pharmaceutical, MSD, ALX Oncology, Debiopharm International, Merck, ISA Pharmaceuticals, Roche Pharmaceuticals, Boehringer Ingelheim, Seagen, Gilead; and travel accommodation expenses from MSD, Merck. J.M.P. declares consulting or advisory roles for Janssen Oncology, Astellas Pharma, VCN Biosciences, Clovis Oncology, Roche/Genentech, Bristol-Myers Squibb, Merck Sharp & Dohme, BeiGene; research funding from Bristol-Myers Squibb, AstraZeneca/MedImmune, Merck Sharp & Dohme, Pfizer/EMD Serono, Incyte, Janssen Oncology; and travel, accommodation and other expenses from Janssen Oncology, Roche, Bristol-Myers Squibb. J.M.-L. has received lecturing fees from Astellas, Bristol-Myers Squibb, MSD, Novartis, Pierre Fabre, Pfizer, Roche, and Sanofi; advisory fees from Bristol-Myers Squibb, Highlight Therapeutics, Novartis, Pierre Fabre, Roche, Sanofi; and travel grants from Bristol-Myers Squibb, Merck, MSD, Novartis, Pierre Fabre, Pfizer, Roche, Ipsen. All other authors declare no potential competing interests.
Figures


References
-
- Cherpelis BS, Marcusen C, Lang PG. Prognostic factors for metastasis in squamous cell carcinoma of the skin. Dermatol. Surg. 2002;28:268–273. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

