Amino acid influx via LAT1 regulates iron demand and sensitivity to PPMX-T003 of aggressive natural killer cell leukemia
- PMID: 38914715
- PMCID: PMC11286515
- DOI: 10.1038/s41375-024-02296-6
Amino acid influx via LAT1 regulates iron demand and sensitivity to PPMX-T003 of aggressive natural killer cell leukemia
Abstract
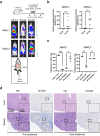
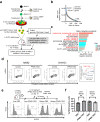
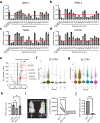
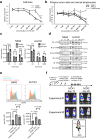
Aggressive natural killer cell leukemia (ANKL) is a rare hematological malignancy with a fulminant clinical course. Our previous study revealed that ANKL cells proliferate predominantly in the liver sinusoids and strongly depend on transferrin supplementation. In addition, we demonstrated that liver-resident ANKL cells are sensitive to PPMX-T003, an anti-human transferrin receptor 1 inhibitory antibody, whereas spleen-resident ANKL cells are resistant to transferrin receptor 1 inhibition. However, the microenvironmental factors that regulate the iron dependency of ANKL cells remain unclear. In this study, we first revealed that the anti-neoplastic effect of PPMX-T003 was characterized by DNA double-strand breaks in a DNA replication-dependent manner, similar to conventional cytotoxic agents. We also found that the influx of extracellular amino acids via LAT1 stimulated sensitivity to PPMX-T003. Taken together, we discovered that the amount of extracellular amino acid influx through LAT1 was the key environmental factor determining the iron dependency of ANKL cells via adjustment of their mTOR/Myc activity, which provides a good explanation for the different sensitivity to PPMX-T003 between liver- and spleen-resident ANKL cells, as the liver sinusoid contains abundant amino acids absorbed from the gut.
© 2024. The Author(s).
Conflict of interest statement
TM was a chief technology officer and an executive officer, and KI was a research manager of Perseus Proteomics Inc. Both has stock options. The authors do not have any other conflict of interest to be disclosed.
Figures

References
MeSH terms
Substances
Grants and funding
- JP23H02940/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP24K19229/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP23K14629/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP22H04922/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22ama221213h0002/Japan Agency for Medical Research and Development (AMED)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

