This is a preprint.
Transport and Organization of Individual Vimentin Filaments Within Dense Networks Revealed by Single Particle Tracking and 3D FIB-SEM
- PMID: 38915582
- PMCID: PMC11195130
- DOI: 10.1101/2024.06.10.598346
Transport and Organization of Individual Vimentin Filaments Within Dense Networks Revealed by Single Particle Tracking and 3D FIB-SEM
Update in
-
Vimentin filament transport and organization revealed by single-particle tracking and 3D FIB-SEM.J Cell Biol. 2025 Apr 7;224(4):e202406054. doi: 10.1083/jcb.202406054. Epub 2025 Mar 10. J Cell Biol. 2025. PMID: 40062969
Abstract
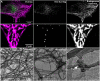
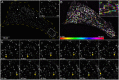
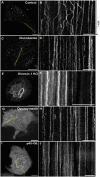
Single-particle tracking demonstrates that individual filaments in bundles of vimentin intermediate filaments are transported in the cytoplasm by motor proteins along microtubules. Furthermore, using 3D FIB-SEM the authors showed that vimentin filament bundles are loosely packed and coaligned with microtubules. Vimentin intermediate filaments (VIFs) form complex, tight-packed networks; due to this density, traditional ensemble labeling and imaging approaches cannot accurately discern single filament behavior. To address this, we introduce a sparse vimentin-SunTag labeling strategy to unambiguously visualize individual filament dynamics. This technique confirmed known long-range dynein and kinesin transport of peripheral VIFs and uncovered extensive bidirectional VIF motion within the perinuclear vimentin network, a region we had thought too densely bundled to permit such motility. To examine the nanoscale organization of perinuclear vimentin, we acquired high-resolution electron microscopy volumes of a vitreously frozen cell and reconstructed VIFs and microtubules within a ~50 μm3 window. Of 583 VIFs identified, most were integrated into long, semi-coherent bundles that fluctuated in width and filament packing density. Unexpectedly, VIFs displayed minimal local co-alignment with microtubules, save for sporadic cross-over sites that we predict facilitate cytoskeletal crosstalk. Overall, this work demonstrates single VIF dynamics and organization in the cellular milieu for the first time.
Conflict of interest statement
Statements and Declarations All authors declare no conflict of interest.
Figures



References
-
- Cheng F., Shen Y., Mohanasundaram P., Lindström M., Ivaska J., Ny T., and Erikss J.E.. 2016. Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-β-Slug signaling. Proc Natl Acad Sci U S A. 113:E4320–E4327. doi:10.1073/pnas.1519197113. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
