This is a preprint.
A mechanism for telomere-specific telomere length regulation
- PMID: 38915611
- PMCID: PMC11195199
- DOI: 10.1101/2024.06.12.598646
A mechanism for telomere-specific telomere length regulation
Abstract
Telomeric DNA, composed of short, direct repeats, is of crucial importance for chromosome stability. Due to intrinsic problems with replicating this DNA, the repeat tracts shorten at each cell division. Once repeat tracts become critically short, a telomeric stress signal induces cellular senescence and division arrest, which eventually may lead to devastating age-related degenerative diseases associated with dysfunctional telomers. Conversely, maintenance of telomere length by telomerase upregulation is a hallmark of cancer. Therefore, telomere length is a critical determinant of telomere function. How telomere length is established and molecular mechanisms for telomere-specific length regulation remained unknown. Here we show that subtelomeric chromatin is a determinant for how telomere equilibrium set-length is established in cis. The results demonstrate that telomerase recruitment mediated by the telomere-associated Sir4 protein is modulated on chromosome 3L in a telomere-specific way. Increased Sir4 abundance on subtelomeric heterochromatin of this specific telomere leads to telomere lengthening of only that telomere in cis, but not at other telomeres. Therefore, this work describes a mechanism for a how telomere-specific repeat tract length can be established. Further, our results will force the evaluation of telomere length away from a generalized view to a more telomere-specific consideration.
Keywords: Ku-proteins; Telomerase RNA; Telomerase recruitment; Telomere length maintenance; Telomeric chromatin.
Conflict of interest statement
CONFLICT OF INTEREST The authors declare no conflict of interest.
Figures
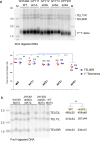




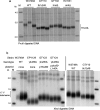

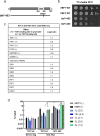
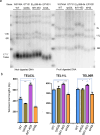


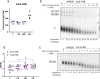

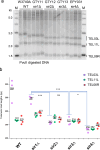
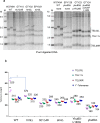
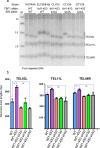
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
