This is a preprint.
Identification and characterization of early human photoreceptor states and cell-state-specific retinoblastoma-related features
- PMID: 38915659
- PMCID: PMC11195049
- DOI: 10.1101/2023.02.28.530247
Identification and characterization of early human photoreceptor states and cell-state-specific retinoblastoma-related features
Update in
-
Identification and characterization of early human photoreceptor states and cell-state-specific retinoblastoma-related features.Elife. 2025 Aug 6;13:RP101918. doi: 10.7554/eLife.101918. Elife. 2025. PMID: 40767624 Free PMC article.
Abstract
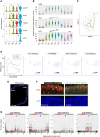
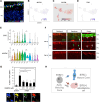
Human cone photoreceptors differ from rods and serve as the retinoblastoma cell-of-origin, yet the developmental basis for their distinct behaviors is poorly understood. Here, we used deep full-length single-cell RNA-sequencing (scRNA-seq) to distinguish post-mitotic cone and rod developmental states and identify cone-specific features that contribute to retinoblastomagenesis. The analyses revealed nascent, immediately post-mitotic cone- and rod precursor populations characterized by higher THRB or NRL regulon activities, immature and maturing cone and rod precursors with concurrent cone- and rod-related gene and regulon expression, and distinct early and late cone and rod maturation states distinguished by maturation-associated declines in RAX regulon activity. Cell-state-specific gene expression features inferred based on full-length scRNA-seq were consistent with past single nucleus 3' RNA-seq analyses. Beyond the cell state characterizations, full-length scRNAseq revealed that both L/M cone and rod precursors co-expressed NRL and THRB RNAs, yet they differentially expressed functionally antagonistic NRL and THRB isoforms and prematurely terminated THRB transcripts. Moreover, early L/M cone precursors exhibited successive expression of several lncRNAs along with MYCN, which composed the seventh most L/M-cone-specific regulon, and SYK, which was implicated in the cone precursors' proliferative response to RB1 loss. These findings reveal previously unresolved photoreceptor precursor states and suggest a role for early cone-precursor-intrinsic SYK expression in retinoblastoma initiation.
Figures








References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
