This is a preprint.
Human cytomegalovirus induces neuronal gene expression for viral maturation
- PMID: 38915666
- PMCID: PMC11195207
- DOI: 10.1101/2024.06.13.598910
Human cytomegalovirus induces neuronal gene expression for viral maturation
Update in
-
Human cytomegalovirus induces neuronal gene expression through IE1 for viral maturation.Nat Commun. 2025 Aug 8;16(1):7316. doi: 10.1038/s41467-025-61915-7. Nat Commun. 2025. PMID: 40781068 Free PMC article.
Abstract
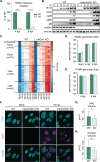
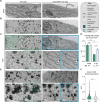
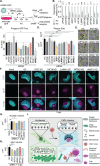
Viral invasion of the host cell causes some of the most dramatic changes in biology. Human cytomegalovirus (HCMV) extensively remodels host cells, altering nuclear shape and generating a cytoplasmic viral-induced assembly compartment (vIAC). How these striking morphology changes take place in the context of host gene regulation is still emerging. Here, we discovered that histone variant macroH2A1 is essential for producing infectious progeny. Because virion maturation and cellular remodeling are closely linked processes, we investigated structural changes in the host cell upon HCMV infection. We discovered that macroH2A1 is necessary for HCMV-induced reorganization of the host nucleus, cytoskeleton, and endoplasmic reticulum. Furthermore, using RNA-seq we found that while all viral genes were highly expressed in the absence of macroH2A1, many HCMV-induced host genes were not. Remarkably, hundreds of these HCMV-induced macroH2A1-dependent host genes are associated with neuronal synapse formation and vesicle trafficking. Knock-down of these HCMV-induced neuronal genes during infection resulted in malformed vIACs and smaller plaques, establishing their importance to HCMV infection. Together, our findings demonstrate that HCMV manipulates host gene expression by hijacking a dormant neuronal secretory pathway for efficient virion maturation.
Figures

References
-
- Cannon M. J., Schmid D. S. & Hyde T. B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Méd. Virol. 20, 202–213 (2010). - PubMed
-
- Fowler K., Mucha J., Neumann M., Lewandowski W., Kaczanowska M., Grys M., Schmidt E., Natenshon A., Talarico C., Buck P. O. & Diaz-Decaro J. A systematic literature review of the global seroprevalence of cytomegalovirus: possible implications for treatment, screening, and vaccine development. BMC Public Heal. 22, 1659 (2022). - PMC - PubMed
-
- Zanghellini F., Boppana S. B., Emery V. C., Griffiths P. D. & Pass R. F. Asymptomatic Primary Cytomegalovirus Infection: Virologic and Immunologic Features. J. Infect. Dis. 180, 702–707 (1999). - PubMed
-
- Azevedo L. S., Pierrotti L. C., Abdala E., Costa S. F., Strabelli T. M. V., Campos S. V., Ramos J. F., Latif A. Z. A., Litvinov N., Maluf N. Z., Filho H. H. C., Pannuti C. S., Lopes M. H., Santos V. A., dos, Linardi C. da C. G., Yasuda. & Marques H. H. de S. Cytomegalovirus infection in transplant recipients. Clinics 70, 515–523 (2015). - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
