Multicountry Spread of Influenza A(H1N1)pdm09 Viruses with Reduced Oseltamivir Inhibition, May 2023-February 2024
- PMID: 38916572
- PMCID: PMC11210663
- DOI: 10.3201/eid3007.240480
Multicountry Spread of Influenza A(H1N1)pdm09 Viruses with Reduced Oseltamivir Inhibition, May 2023-February 2024
Abstract
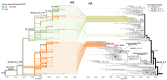
Since May 2023, a novel combination of neuraminidase mutations, I223V + S247N, has been detected in influenza A(H1N1)pdm09 viruses collected in countries spanning 5 continents, mostly in Europe (67/101). The viruses belong to 2 phylogenetically distinct groups and display ≈13-fold reduced inhibition by oseltamivir while retaining normal susceptibility to other antiviral drugs.
Keywords: H1N1; Influenza; United States; antimicrobial resistance; antiviral; baloxavir; influenza A(H1N1)pdm09; neuraminidase inhibitors; oseltamivir; pH1N1; phenotypic testing; reduced inhibition; substitution; viruses.
Figures

References
-
- Centers for Disease Control and Prevention. Influenza antiviral drug resistance [cited 2024 Mar 18]. https://www.cdc.gov/flu/treatment/antiviralresistance.htm
-
- World Health Organization-Antiviral Working Group. Summary of neuraminidase (NA) amino acid substitutions assessed for their effects on inhibition by neuraminidase inhibitors (NAIs). 2023. Mar 7 [cited 2024 Mar 18]. https://cdn.who.int/media/docs/default-source/global-influenza-programme...
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

