Pathogenic variants in autism gene KATNAL2 cause hydrocephalus and disrupt neuronal connectivity by impairing ciliary microtubule dynamics
- PMID: 38916997
- PMCID: PMC11228466
- DOI: 10.1073/pnas.2314702121
Pathogenic variants in autism gene KATNAL2 cause hydrocephalus and disrupt neuronal connectivity by impairing ciliary microtubule dynamics
Abstract
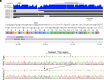
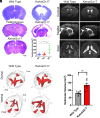
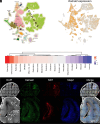
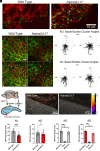
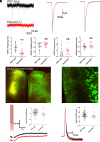
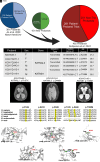
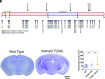
Enlargement of the cerebrospinal fluid (CSF)-filled brain ventricles (cerebral ventriculomegaly), the cardinal feature of congenital hydrocephalus (CH), is increasingly recognized among patients with autism spectrum disorders (ASD). KATNAL2, a member of Katanin family microtubule-severing ATPases, is a known ASD risk gene, but its roles in human brain development remain unclear. Here, we show that nonsense truncation of Katnal2 (Katnal2Δ17) in mice results in classic ciliopathy phenotypes, including impaired spermatogenesis and cerebral ventriculomegaly. In both humans and mice, KATNAL2 is highly expressed in ciliated radial glia of the fetal ventricular-subventricular zone as well as in their postnatal ependymal and neuronal progeny. The ventriculomegaly observed in Katnal2Δ17 mice is associated with disrupted primary cilia and ependymal planar cell polarity that results in impaired cilia-generated CSF flow. Further, prefrontal pyramidal neurons in ventriculomegalic Katnal2Δ17 mice exhibit decreased excitatory drive and reduced high-frequency firing. Consistent with these findings in mice, we identified rare, damaging heterozygous germline variants in KATNAL2 in five unrelated patients with neurosurgically treated CH and comorbid ASD or other neurodevelopmental disorders. Mice engineered with the orthologous ASD-associated KATNAL2 F244L missense variant recapitulated the ventriculomegaly found in human patients. Together, these data suggest KATNAL2 pathogenic variants alter intraventricular CSF homeostasis and parenchymal neuronal connectivity by disrupting microtubule dynamics in fetal radial glia and their postnatal ependymal and neuronal descendants. The results identify a molecular mechanism underlying the development of ventriculomegaly in a genetic subset of patients with ASD and may explain persistence of neurodevelopmental phenotypes in some patients with CH despite neurosurgical CSF shunting.
Keywords: autism; cerebrospinal fluid dynamics; ciliopathy; hydrocephalus; structural brain disorder.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
Comment in
-
KATNAL2 mutations link ciliary dysfunction to hydrocephalus and autism.Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2410761121. doi: 10.1073/pnas.2410761121. Epub 2024 Jul 15. Proc Natl Acad Sci U S A. 2024. PMID: 39008680 Free PMC article. No abstract available.
Similar articles
-
Loss of Katnal2 leads to ependymal ciliary hyperfunction and autism-related phenotypes in mice.PLoS Biol. 2024 May 8;22(5):e3002596. doi: 10.1371/journal.pbio.3002596. eCollection 2024 May. PLoS Biol. 2024. PMID: 38718086 Free PMC article.
-
Disruption of the mouse Jhy gene causes abnormal ciliary microtubule patterning and juvenile hydrocephalus.Dev Biol. 2013 Oct 1;382(1):172-85. doi: 10.1016/j.ydbio.2013.07.003. Epub 2013 Jul 29. Dev Biol. 2013. PMID: 23906841 Free PMC article.
-
Daple Coordinates Planar Polarized Microtubule Dynamics in Ependymal Cells and Contributes to Hydrocephalus.Cell Rep. 2017 Jul 25;20(4):960-972. doi: 10.1016/j.celrep.2017.06.089. Cell Rep. 2017. PMID: 28746879
-
Rethinking the cilia hypothesis of hydrocephalus.Neurobiol Dis. 2022 Dec;175:105913. doi: 10.1016/j.nbd.2022.105913. Epub 2022 Oct 29. Neurobiol Dis. 2022. PMID: 36341771 Review.
-
Exploring mechanisms of ventricular enlargement in idiopathic normal pressure hydrocephalus: a role of cerebrospinal fluid dynamics and motile cilia.Fluids Barriers CNS. 2021 Apr 19;18(1):20. doi: 10.1186/s12987-021-00243-6. Fluids Barriers CNS. 2021. PMID: 33874972 Free PMC article. Review.
Cited by
-
Circadian cilia transcriptome in mouse brain across physiological and pathological states.Mol Brain. 2024 Sep 20;17(1):67. doi: 10.1186/s13041-024-01143-0. Mol Brain. 2024. PMID: 39304885 Free PMC article.
-
KATNAL2 mutations link ciliary dysfunction to hydrocephalus and autism.Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2410761121. doi: 10.1073/pnas.2410761121. Epub 2024 Jul 15. Proc Natl Acad Sci U S A. 2024. PMID: 39008680 Free PMC article. No abstract available.
References
-
- Kahle K. T., Kulkarni A. V., Limbrick D. D. Jr., Warf B. C., Hydrocephalus in children. Lancet 387, 788–799 (2016). - PubMed
-
- Svancer P., Spaniel F., Brain ventricular volume changes in schizophrenia. A narrative review. Neurosci. Lett. 759, 136065 (2021). - PubMed
-
- Riva-Cambrin J., et al. , Impact of ventricle size on neuropsychological outcomes in treated pediatric hydrocephalus: An HCRN prospective cohort study. J. Neurosurg. Pediatr. 29, 245–256 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- R01 NS109358/NS/NINDS NIH HHS/United States
- 1R01NS109358-01/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R01 NS111029/NS/NINDS NIH HHS/United States
- P30 CA023108/CA/NCI NIH HHS/United States
- R01NS127879/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- T32 GM007205/GM/NIGMS NIH HHS/United States
- 1R01NS111029-01A1/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 MH124934/MH/NIMH NIH HHS/United States
- F31 NS115519/NS/NINDS NIH HHS/United States
- R01 MH097949/MH/NIMH NIH HHS/United States
- F31NS115519/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 NS127879/NS/NINDS NIH HHS/United States
- R01 NS131610/NS/NINDS NIH HHS/United States
- 1R01MH097949/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

