MRNIP limits ssDNA gaps during replication stress
- PMID: 38917325
- PMCID: PMC11317133
- DOI: 10.1093/nar/gkae546
MRNIP limits ssDNA gaps during replication stress
Abstract
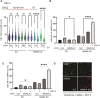
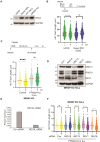
Replication repriming by the specialized primase-polymerase PRIMPOL ensures the continuity of DNA synthesis during replication stress. PRIMPOL activity generates residual post-replicative single-stranded nascent DNA gaps, which are linked with mutagenesis and chemosensitivity in BRCA1/2-deficient models, and which are suppressed by replication fork reversal mediated by the DNA translocases SMARCAL1 and ZRANB3. Here, we report that the MRE11 regulator MRNIP limits the prevalence of PRIMPOL and MRE11-dependent ssDNA gaps in cells in which fork reversal is perturbed either by treatment with the PARP inhibitor Olaparib, or by depletion of SMARCAL1 or ZRANB3. MRNIP-deficient cells are sensitive to PARP inhibition and accumulate PRIMPOL-dependent DNA damage, supportive of a pro-survival role for MRNIP linked to the regulation of gap prevalence. In MRNIP-deficient cells, post-replicative gap filling is driven in S-phase by UBC13-mediated template switching involving REV1 and the TLS polymerase Pol-ζ. Our findings represent the first report of modulation of post-replicative ssDNA gap dynamics by a direct MRE11 regulator.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

