Chemokine-mediated cell migration into the central nervous system in progressive multifocal leukoencephalopathy
- PMID: 38917802
- PMCID: PMC11293326
- DOI: 10.1016/j.xcrm.2024.101622
Chemokine-mediated cell migration into the central nervous system in progressive multifocal leukoencephalopathy
Abstract
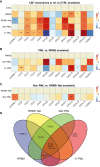
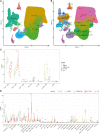
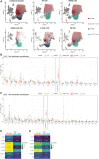
Progressive multifocal leukoencephalopathy (PML) has been associated with different forms of immune compromise. This study analyzes the chemokine signals and attracted immune cells in cerebrospinal fluid (CSF) during PML to define immune cell subpopulations relevant for the PML immune response. In addition to chemokines that indicate a general state of inflammation, like CCL5 and CXCL10, the CSF of PML patients specifically contains CCL2 and CCL4. Single-cell transcriptomics of CSF cells suggests an enrichment of distinct CD4+ and CD8+ T cells expressing chemokine receptors CCR2, CCR5, and CXCR3, in addition to ITGA4 and the genetic PML risk genes STXBP2 and LY9. This suggests that specific immune cell subpopulations migrate into the central nervous system to mitigate PML, and their absence might coincide with PML development. Monitoring them might hold clues for PML risk, and boosting their recruitment or function before therapeutic immune reconstitution might improve its risk-benefit ratio.
Keywords: HIV; central nervous system; cerebrospinal fluid; chemokines; immune cell migration; multiple sclerosis; natalizumab; progressive multifocal leukoencephalopathy; single-cell RNA sequencing; viral encephalitis.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.M.-M. reported travel grants from Alexion outside of the submitted work. S.G. was a consultant for Biogen during the initial phases of the study. She is the scientific director for the Myelin Repair Foundation. E.C.-M. was an employee of Biogen during the conduct of the study. G.M.z.H. reported speaker honoraria and consultant/advisor reimbursement from Roche, LFB Pharma, and Alexion. L.K. reported personal fees from Alexion, Bayer, Biogen, Celgene, Sanofi, Horizon, Grifols, Merck Serono, Novartis, Roche, Santhera, and Teva and grants from the German Research Foundation, IZKF Münster, IMF Münster, Biogen, Immunic AG, Novartis, and Merck Serono outside the submitted work. C.C.G. reported grants from DFG SFB/TR128 A09 during the conduct of the study; grants from DFG (single grant GR3946-3/1), IZKF (grant Kl13_010_19), Horizon2020 ReSToRe, Biogen, Roche, and Novartis Pharma; personal fees from MyLan and DIU Dresden International University GmbH; and other from Biogen, Euroimmun, MyLan, and Novartis Pharma outside the submitted work. H.W. reported personal fees from AbbVie, Alexion, Argenx, Biogen, Bristol-Myers Squibb/Celgene, EMD Serono, F. Hoffmann-La Roche Ltd., Fondazione Cariplo, Genzyme, Gossamer Bio, Idorsia, Immunic, Immunovant, Janssen, Lundbeck, Merck, Neurodiem, NexGen, Novartis, PSI CRO, Roche Pharma AG, Sanofi, Swiss Multiple Sclerosis Society TEVA, UCB Biopharma, WebMD Global, and Worldwide Clinical Trials outside the submitted work. He reported grants by the DFG (CRC128 A09 and 445569437) during the conduct of the study and funding by the German Federal Ministry for Education and Research (BMBF), Deutsche Myasthenie Gesellschaft e.V., Alexion, Amicus Therapeutics Inc., Argenx, Biogen, CSL Behring, Roche, Genzyme, Merck, Novartis Pharma, Roche Pharma, and UCB Biopharma outside of the submitted work. N.S. reported grants from DFG and Biogen during the conduct of the study.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

