Tracking the Spread of the BA.2.86 Lineage in Italy Through Wastewater Analysis
- PMID: 38918335
- PMCID: PMC11525314
- DOI: 10.1007/s12560-024-09607-1
Tracking the Spread of the BA.2.86 Lineage in Italy Through Wastewater Analysis
Abstract
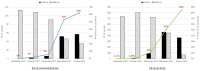
The emergence of new SARS-CoV-2 variants poses challenges to global surveillance efforts, necessitating swift actions in their detection, evaluation, and management. Among the most recent variants, Omicron BA.2.86 and its sub-lineages have gained attention due to their potential immune evasion properties. This study describes the development of a digital PCR assay for the rapid detection of BA.2.86 and its descendant lineages, in wastewater samples. By using this assay, we analyzed wastewater samples collected in Italy from September 2023 to January 2024. Our analysis revealed the presence of BA.2.86 lineages already in October 2023 with a minimal detection rate of 2% which then rapidly increased, becoming dominant by January 2024, accounting for a prevalence of 62%. The findings emphasize the significance of wastewater-based surveillance in tracking emerging variants and underscore the efficacy of targeted digital PCR assays for environmental monitoring.
Keywords: BA.2.86; Digital PCR; JN.1; SARS-CoV-2; Wastewater surveillance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Attar Cohen, H., Mesfin, S., Ikejezie, J., Kassamali, Z., Campbell, F., Adele, S., Guinko, N., Idoko, F., Mirembe, B. B., Mitri, M. E., Nezu, I., Shimizu, K., Ngongheh, A. B., Sklenovska, N., Gumede, N., Mosha, F. S., Mohamed, B., Corpuz, A., Pebody, R., Pavlin, B. I. (2023). Surveillance for variants of SARS-CoV-2 to inform risk assessments. Bulletin of the World Health Organization,101(11), 707–716. 10.2471/BLT.23.290093 - PMC - PubMed
-
- Bartel, A., Grau, J. H., Bitzegeio, J., Werber, D., Linzner, N., Schumacher, V., Garske, S., Liere, K., Hackenbeck, T., Rupp, S. I., Sagebiel, D., Böckelmann, U., & Meixner, M. (2024). Timely monitoring of SARS-CoV-2 RNA fragments in wastewater shows the emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany. Viruses,16(1), 102. 10.3390/v16010102 - PMC - PubMed
-
- Bonanno Ferraro, G., Veneri, C., Mancini, P., Iaconelli, M., Suffredini, E., Bonadonna, L., Lucentini, L., Bowo-Ngandji, A., Kengne-Nde, C., Mbaga, D. S., Mahamat, G., Tazokong, H. R., Ebogo-Belobo, J. T., Njouom, R., Kenmoe, S., La Rosa, G. (2022). A State-of-the-Art scoping review on SARS-CoV-2 in sewage focusing on the potential of wastewater surveillance for the monitoring of the COVID-19 pandemic. Food and Environmental Virology,14(4), 315–354. 10.1007/s12560-021-09498-6 - PMC - PubMed
-
- Caccuri, F., Messali, S., Scarpa, F., Giovanetti, M., Ciccozzi, M., & Caruso, A. (2023). First detection of SARS-CoV-2 BA.2.86.1 in Italy. Journal of Medical Virology,95(10), 29175. 10.1002/jmv.29175 - PubMed
-
- Combe, M., Cherif, E., Deremarque, T., Rivera-Ingraham, G., Seck-Thiam, F., Justy, F., Doudou, J. C., Carod, J. F., Carage, T., Procureur, A., & Gozlan, R. E. (2024). Wastewater sequencing as a powerful tool to reveal SARS-CoV-2 variant introduction and spread in French Guiana, South America. The Science of the Total Environment,924, 171645. 10.1016/j.scitotenv.2024.171645 - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

