SDS22 coordinates the assembly of holoenzymes from nascent protein phosphatase-1
- PMID: 38918402
- PMCID: PMC11199634
- DOI: 10.1038/s41467-024-49746-4
SDS22 coordinates the assembly of holoenzymes from nascent protein phosphatase-1
Abstract
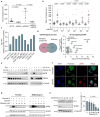
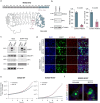
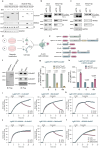
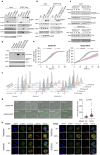
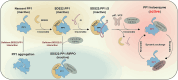
SDS22 forms an inactive complex with nascent protein phosphatase PP1 and Inhibitor-3. SDS22:PP1:Inhibitor-3 is a substrate for the ATPase p97/VCP, which liberates PP1 for binding to canonical regulatory subunits. The exact role of SDS22 in PP1-holoenzyme assembly remains elusive. Here, we show that SDS22 stabilizes nascent PP1. In the absence of SDS22, PP1 is gradually lost, resulting in substrate hyperphosphorylation and a proliferation arrest. Similarly, we identify a female individual with a severe neurodevelopmental disorder bearing an unstable SDS22 mutant, associated with decreased PP1 levels. We furthermore find that SDS22 directly binds to Inhibitor-3 and that this is essential for the stable assembly of SDS22:PP1: Inhibitor-3, the recruitment of p97/VCP, and the extraction of SDS22 during holoenzyme assembly. SDS22 with a disabled Inhibitor-3 binding site co-transfers with PP1 to canonical regulatory subunits, thereby forming non-functional holoenzymes. Our data show that SDS22, through simultaneous interaction with PP1 and Inhibitor-3, integrates the major steps of PP1 holoenzyme assembly.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

