Transcription regulates the spatio-temporal dynamics of genes through micro-compartmentalization
- PMID: 38918438
- PMCID: PMC11199603
- DOI: 10.1038/s41467-024-49727-7
Transcription regulates the spatio-temporal dynamics of genes through micro-compartmentalization
Abstract
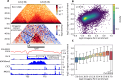
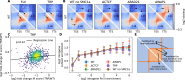
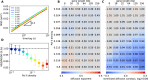
Although our understanding of the involvement of heterochromatin architectural factors in shaping nuclear organization is improving, there is still ongoing debate regarding the role of active genes in this process. In this study, we utilize publicly-available Micro-C data from mouse embryonic stem cells to investigate the relationship between gene transcription and 3D gene folding. Our analysis uncovers a nonmonotonic - globally positive - correlation between intragenic contact density and Pol II occupancy, independent of cohesin-based loop extrusion. Through the development of a biophysical model integrating the role of transcription dynamics within a polymer model of chromosome organization, we demonstrate that Pol II-mediated attractive interactions with limited valency between transcribed regions yield quantitative predictions consistent with chromosome-conformation-capture and live-imaging experiments. Our work provides compelling evidence that transcriptional activity shapes the 4D genome through Pol II-mediated micro-compartmentalization.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

