Parallel degradome-seq and DMS-MaPseq substantially revise the miRNA biogenesis atlas in Arabidopsis
- PMID: 38918606
- PMCID: PMC11578046
- DOI: 10.1038/s41477-024-01725-9
Parallel degradome-seq and DMS-MaPseq substantially revise the miRNA biogenesis atlas in Arabidopsis
Abstract
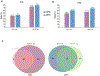
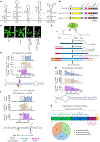
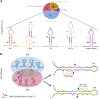
MicroRNAs (miRNAs) are produced from highly structured primary transcripts (pri-miRNAs) and regulate numerous biological processes in eukaryotes. Due to the extreme heterogeneity of these structures, the initial processing sites of plant pri-miRNAs and the structural rules that determine their processing have been predicted for many miRNAs but remain elusive for others. Here we used semi-active DCL1 mutants and advanced degradome-sequencing strategies to accurately identify the initial processing sites for 147 of 326 previously annotated Arabidopsis miRNAs and to illustrate their associated pri-miRNA cleavage patterns. Elucidating the in vivo RNA secondary structures of 73 pri-miRNAs revealed that about 95% of them differ from in silico predictions, and that the revised structures offer clearer interpretation of the processing sites and patterns. Finally, DCL1 partners Serrate and HYL1 could synergistically and independently impact processing patterns and in vivo RNA secondary structures of pri-miRNAs. Together, our work sheds light on the precise processing mechanisms of plant pri-miRNAs.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures
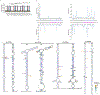













Comment in
-
Exploring the landscape of miRNA production and the structural rules that shape it.Nat Plants. 2024 Jul;10(7):1060-1061. doi: 10.1038/s41477-024-01728-6. Nat Plants. 2024. PMID: 38956418 No abstract available.
References
-
- Song XW, Li Y, Cao XF & Qi YJ MicroRNAs and their regulatory roles in plant–environment interactions. Annu. Rev. Plant Biol 70, 489–525 (2019). - PubMed
-
- Li S, Castillo-González C, Yu B & Zhang X The functions of plant small RNA s in development and in stress responses. Plant J. 90, 654–670 (2017). - PubMed
-
- Ma Z & Zhang X Actions of plant Argonautes: predictable or unpredictable? Curr. Opin. Plant Biol 45, 59–67 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

