Maternal SARS-CoV-2 impacts fetal placental macrophage programs and placenta-derived microglial models of neurodevelopment
- PMID: 38918792
- PMCID: PMC11197235
- DOI: 10.1186/s12974-024-03157-w
Maternal SARS-CoV-2 impacts fetal placental macrophage programs and placenta-derived microglial models of neurodevelopment
Abstract
Background: The SARS-CoV-2 virus activates maternal and placental immune responses. Such activation in the setting of other infections during pregnancy is known to impact fetal brain development. The effects of maternal immune activation on neurodevelopment are mediated at least in part by fetal brain microglia. However, microglia are inaccessible for direct analysis, and there are no validated non-invasive surrogate models to evaluate in utero microglial priming and function. We have previously demonstrated shared transcriptional programs between microglia and Hofbauer cells (HBCs, or fetal placental macrophages) in mouse models.
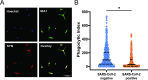
Methods and results: We assessed the impact of maternal SARS-CoV-2 on HBCs isolated from 24 term placentas (N = 10 SARS-CoV-2 positive cases, 14 negative controls). Using single-cell RNA-sequencing, we demonstrated that HBC subpopulations exhibit distinct cellular programs, with specific subpopulations differentially impacted by SARS-CoV-2. Assessment of differentially expressed genes implied impaired phagocytosis, a key function of both HBCs and microglia, in some subclusters. Leveraging previously validated models of microglial synaptic pruning, we showed that HBCs isolated from placentas of SARS-CoV-2 positive pregnancies can be transdifferentiated into microglia-like cells (HBC-iMGs), with impaired synaptic pruning behavior compared to HBC models from negative controls.
Conclusion: These findings suggest that HBCs isolated at birth can be used to create personalized cellular models of offspring microglial programming.
Keywords: COVID-19; Fetal brain; Hofbauer cells; Microglia; Neurodevelopment; Neuroimmune; Placenta; SARS-CoV-2; Single-cell RNA sequencing.
© 2024. The Author(s).
Conflict of interest statement
R.H.P. has received fees for service as a scientific advisor to Belle Artificial Intelligence, Burrage Capital, Circular Genomics, Genomind, Swan Artificial Intelligence Studios, and Vault Health. A.G.E. serves as a consultant for Mirvie, Inc. and receives research funding from Merck Pharmaceuticals outside of this work. The other authors have no disclosures to report.
Figures




Update of
-
Maternal SARS-CoV-2 impacts fetal placental macrophage programs and placenta-derived microglial models of neurodevelopment.medRxiv [Preprint]. 2023 Dec 30:2023.12.29.23300544. doi: 10.1101/2023.12.29.23300544. medRxiv. 2023. Update in: J Neuroinflammation. 2024 Jun 25;21(1):163. doi: 10.1186/s12974-024-03157-w. PMID: 38234776 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

