Revealing the genetic complexity of hypothyroidism: integrating complementary association methods
- PMID: 38919955
- PMCID: PMC11196612
- DOI: 10.3389/fgene.2024.1409226
Revealing the genetic complexity of hypothyroidism: integrating complementary association methods
Abstract
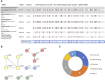
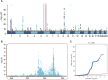
Hypothyroidism is a common endocrine disorder whose prevalence increases with age. The disease manifests itself when the thyroid gland fails to produce sufficient thyroid hormones. The disorder includes cases of congenital hypothyroidism (CH), but most cases exhibit hormonal feedback dysregulation and destruction of the thyroid gland by autoantibodies. In this study, we sought to identify causal genes for hypothyroidism in large populations. The study used the UK-Biobank (UKB) database, reporting on 13,687 cases of European ancestry. We used GWAS compilation from Open Targets (OT) and tuned protocols focusing on genes and coding regions, along with complementary association methods of PWAS (proteome-based) and TWAS (transcriptome-based). Comparing summary statistics from numerous GWAS revealed a limited number of variants associated with thyroid development. The proteome-wide association study method identified 77 statistically significant genes, half of which are located within the Chr6-MHC locus and are enriched with autoimmunity-related genes. While coding GWAS and PWAS highlighted the centrality of immune-related genes, OT and transcriptome-wide association study mostly identified genes involved in thyroid developmental programs. We used independent populations from Finland (FinnGen) and the Taiwan cohort to validate the PWAS results. The higher prevalence in females relative to males is substantiated as the polygenic risk score prediction of hypothyroidism relied mostly from the female group genetics. Comparing results from OT, TWAS, and PWAS revealed the complementary facets of hypothyroidism's etiology. This study underscores the significance of synthesizing gene-phenotype association methods for this common, intricate disease. We propose that the integration of established association methods enhances interpretability and clinical utility.
Keywords: FinnGen; GWAS; Hashimoto’s thyroiditis; PWAS; UK-Biobank; congenital hypothyroidism; genotyping; open targets.
Copyright © 2024 Zucker, Kovalerchik, Stern, Kaufman and Linial.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Al Taji E., Biebermann H., Límanová Z. k., Hníková O., Zikmund J., Dame C., et al. (2007). Screening for mutations in transcription factors in a Czech cohort of 170 patients with congenital and early-onset hypothyroidism: identification of a novel PAX8 mutation in dominantly inherited early-onset non-autoimmune hypothyroidism. Eur. J. Endocrinol. 156, 521–529. 10.1530/EJE-06-0709 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

