Multi-COBRA hemagglutinin formulated with cGAMP microparticles elicits protective immune responses against influenza viruses
- PMID: 38920382
- PMCID: PMC11288037
- DOI: 10.1128/msphere.00160-24
Multi-COBRA hemagglutinin formulated with cGAMP microparticles elicits protective immune responses against influenza viruses
Abstract
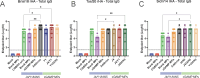
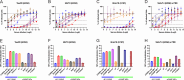
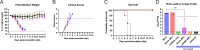
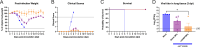
In humans, seasonal influenza viruses cause epidemics. Avian influenza viruses are of particular concern because they can infect multiple species and lead to unpredictable and severe disease. Therefore, there is an urgent need for a universal influenza vaccine that provides protection against all influenza strains. The cyclic GMP-AMP (cGAMP) is a promising adjuvant for subunit vaccines, which promotes type I interferons' production through the stimulator of interferon genes (STING) pathway. The encapsulation of cGAMP in acetalated dextran (Ace-DEX) microparticles (MPs) enhances its intracellular delivery. In this study, the Computationally Optimized Broadly Reactive Antigen (COBRA) methodology was used to generate H1, H3, and H5 vaccine candidates. Monovalent and multivalent COBRA HA vaccines formulated with cGAMP Ace-DEX MPs were evaluated in mice for protective antibody responses. cGAMP MPs adjuvanted COBRA HA vaccines elicited robust antigen-specific antibodies following vaccination. Compared with COBRA HA vaccine groups with no adjuvant or blank MPs, the cGAMP MPs enhanced HAI activity elicited by COBRA HA vaccines. The HAI activity was not significantly different between cGAMP MPs adjuvanted monovalent or multivalent COBRA HA vaccines. The cGAMP MPs adjuvanted COBRA vaccine groups had higher antigen-specific IgG2a-binding titers than the COBRA vaccine groups with no adjuvant or blank MPs. The COBRA vaccines formulated with cGAMP MPs mitigated diseases caused by influenza viral challenge and decreased pulmonary viral titers in mice. Therefore, the formulation of COBRA vaccines plus cGAMP MPs is a promising universal influenza vaccine that elicits protective immune responses against human seasonal and pre-pandemic strains.
Importance: Influenza viruses cause severe respiratory disease, particularly in the very young and the elderly. Next-generation influenza vaccines are needed to protect against new influenza variants. This report used a promising adjuvant, cyclic GMP-AMP (cGAMP), to enhance the elicited antibodies by an improved influenza hemagglutinin candidate and protect against influenza virus infection. Overall, adding adjuvants to influenza vaccines is an effective method to improve vaccines.
Keywords: COBRA; acetalated-dextran (Ace-DEX); cGAMP; influenza; mice; microparticles (MPs); vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Update of
-
Multi-COBRA hemagglutinin formulated with cGAMP microparticles elicit protective immune responses against influenza viruses.bioRxiv [Preprint]. 2024 Feb 29:2024.02.27.582355. doi: 10.1101/2024.02.27.582355. bioRxiv. 2024. Update in: mSphere. 2024 Jul 30;9(7):e0016024. doi: 10.1128/msphere.00160-24. PMID: 38464191 Free PMC article. Updated. Preprint.
References
-
- Paget J, Spreeuwenberg P, Charu V, Taylor RJ, Iuliano AD, Bresee J, Simonsen L, Viboud C, Global Seasonal Influenza-associated Mortality Collaborator Network and GLaMOR Collaborating Teams* . 2019. Global mortality associated with seasonal influenza epidemics: new burden estimates and predictors from the GLaMOR Project. J Glob Health 9:020421. doi:10.7189/jogh.09.020421 - DOI - PMC - PubMed
-
- Smith W, Andrewes CH, Laidlaw PP. 1933. A virus obtained from influenza patients. Lancet 222:66–68. doi:10.1016/S0140-6736(00)78541-2 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
