Multiomics Analysis of the PHLDA Gene Family in Different Cancers and Their Clinical Prognostic Value
- PMID: 38921000
- PMCID: PMC11201736
- DOI: 10.3390/cimb46060328
Multiomics Analysis of the PHLDA Gene Family in Different Cancers and Their Clinical Prognostic Value
Abstract
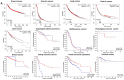
The PHLDA (pleckstrin homology-like domain family) gene family is popularly known as a potential biomarker for cancer identification, and members of the PHLDA family have become considered potentially viable targets for cancer treatments. The PHLDA gene family consists of PHLDA1, PHLDA2, and PHLDA3. The predictive significance of PHLDA genes in cancer remains unclear. To determine the role of pleckstrin as a prognostic biomarker in human cancers, we conducted a systematic multiomics investigation. Through various survival analyses, pleckstrin expression was evaluated, and their predictive significance in human tumors was discovered using a variety of online platforms. By analyzing the protein-protein interactions, we also chose a collection of well-known functional protein partners for pleckstrin. Investigations were also carried out on the relationship between pleckstrins and other cancers regarding mutations and copy number alterations. The cumulative impact of pleckstrin and their associated genes on various cancers, Gene Ontology (GO), and pathway analyses were used for their evaluation. Thus, the expression profiles of PHLDA family members and their prognosis in various cancers may be revealed by this study. During this multiomics analysis, we found that among the PHLDA family, PHLDA1 may be a therapeutic target for several cancers, including kidney, colon, and brain cancer, while PHLDA2 can be a therapeutic target for cancers of the colon, esophagus, and pancreas. Additionally, PHLDA3 may be a useful therapeutic target for ovarian, renal, and gastric cancer.
Keywords: PHLDA (pleckstrin homology-like domain family) gene; cancer biomarker; gene ontology (GO); gene pathway; multiomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
The expression patterns and prognostic significance of pleckstrin homology-like domain family A (PHLDA) in lung cancer and malignant mesothelioma.J Thorac Dis. 2021 Feb;13(2):689-707. doi: 10.21037/jtd-20-2909. J Thorac Dis. 2021. PMID: 33717542 Free PMC article.
-
PHLDA1, another PHLDA family protein that inhibits Akt.Cancer Sci. 2018 Nov;109(11):3532-3542. doi: 10.1111/cas.13796. Epub 2018 Oct 13. Cancer Sci. 2018. PMID: 30207029 Free PMC article.
-
Prognostic Value, Immune Signature, and Molecular Mechanisms of the PHLDA Family in Pancreatic Adenocarcinoma.Int J Mol Sci. 2022 Sep 7;23(18):10316. doi: 10.3390/ijms231810316. Int J Mol Sci. 2022. PMID: 36142223 Free PMC article.
-
Expanding on roles of pleckstrin homology-like domain family A member 1 protein.Cell Tissue Res. 2025 Jan;399(1):9-25. doi: 10.1007/s00441-024-03942-2. Epub 2024 Dec 4. Cell Tissue Res. 2025. PMID: 39630301 Free PMC article. Review.
-
PHLD Class Proteins: A Family of New Players in the p53 Network.Int J Mol Sci. 2020 May 17;21(10):3543. doi: 10.3390/ijms21103543. Int J Mol Sci. 2020. PMID: 32429563 Free PMC article. Review.
References
-
- Baldavira C.M., Machado-Rugolo J., Prieto T.G., Bastos D.R., Balancin M., Ab’Saber A.M., Yaegashi L.B., Souza P.C., Farhat C., Takagaki T.Y., et al. The expression patterns and prognostic significance of pleckstrin homology-like domain family A (PHLDA) in lung cancer and malignant mesothelioma. J. Thorac. Dis. 2021;13:689–707. doi: 10.21037/jtd-20-2909. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

