Characterizing Cellular Physiological States with Three-Dimensional Shape Descriptors for Cell Membranes
- PMID: 38921504
- PMCID: PMC11205511
- DOI: 10.3390/membranes14060137
Characterizing Cellular Physiological States with Three-Dimensional Shape Descriptors for Cell Membranes
Abstract
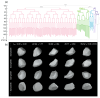
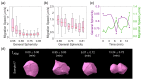
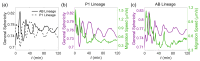
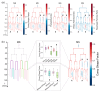
The shape of a cell as defined by its membrane can be closely associated with its physiological state. For example, the irregular shapes of cancerous cells and elongated shapes of neuron cells often reflect specific functions, such as cell motility and cell communication. However, it remains unclear whether and which cell shape descriptors can characterize different cellular physiological states. In this study, 12 geometric shape descriptors for a three-dimensional (3D) object were collected from the previous literature and tested with a public dataset of ~400,000 independent 3D cell regions segmented based on fluorescent labeling of the cell membranes in Caenorhabditis elegans embryos. It is revealed that those shape descriptors can faithfully characterize cellular physiological states, including (1) cell division (cytokinesis), along with an abrupt increase in the elongation ratio; (2) a negative correlation of cell migration speed with cell sphericity; (3) cell lineage specification with symmetrically patterned cell shape changes; and (4) cell fate specification with differential gene expression and differential cell shapes. The descriptors established may be used to identify and predict the diverse physiological states in numerous cells, which could be used for not only studying developmental morphogenesis but also diagnosing human disease (e.g., the rapid detection of abnormal cells).
Keywords: 3D shape descriptor; Caenorhabditis elegans; cell division (cytokinesis); cell fate; cell lineage; cell membrane; cell migration; embryogenesis; fluorescence imaging; gene expression.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Improved 3D cellular morphometry of Caenorhabditis elegans embryos using a refractive index matching medium.PLoS One. 2020 Sep 30;15(9):e0238955. doi: 10.1371/journal.pone.0238955. eCollection 2020. PLoS One. 2020. PMID: 32997668 Free PMC article.
-
Mutations in the Caenorhabditis elegans gene vab-3 reveal distinct roles in fate specification and unequal cytokinesis in an asymmetric cell division.Dev Biol. 1995 Aug;170(2):679-89. doi: 10.1006/dbio.1995.1246. Dev Biol. 1995. PMID: 7649393
-
Systems-level quantification of division timing reveals a common genetic architecture controlling asynchrony and fate asymmetry.Mol Syst Biol. 2015 Jun 10;11(6):814. doi: 10.15252/msb.20145857. Mol Syst Biol. 2015. PMID: 26063786 Free PMC article.
-
Caenorhabditis elegans Gastrulation: A Model for Understanding How Cells Polarize, Change Shape, and Journey Toward the Center of an Embryo.Genetics. 2020 Feb;214(2):265-277. doi: 10.1534/genetics.119.300240. Genetics. 2020. PMID: 32029580 Free PMC article. Review.
-
The Caenorhabditis elegans Excretory System: A Model for Tubulogenesis, Cell Fate Specification, and Plasticity.Genetics. 2016 May;203(1):35-63. doi: 10.1534/genetics.116.189357. Genetics. 2016. PMID: 27183565 Free PMC article. Review.
Cited by
-
Cell lineage-resolved embryonic morphological map reveals signaling associated with cell fate and size asymmetry.Nat Commun. 2025 Apr 18;16(1):3700. doi: 10.1038/s41467-025-58878-0. Nat Commun. 2025. PMID: 40251161 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

