Characterization and Biosynthetic Regulation of Isoflavone Genistein in Deep-Sea Actinomycetes Microbacterium sp. B1075
- PMID: 38921587
- PMCID: PMC11205022
- DOI: 10.3390/md22060276
Characterization and Biosynthetic Regulation of Isoflavone Genistein in Deep-Sea Actinomycetes Microbacterium sp. B1075
Abstract
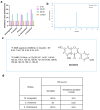
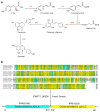
Deep-sea environments, as relatively unexplored extremes within the Earth's biosphere, exhibit notable distinctions from terrestrial habitats. To thrive in these extreme conditions, deep-sea actinomycetes have evolved unique biochemical metabolisms and physiological capabilities to ensure their survival in this niche. In this study, five actinomycetes strains were isolated and identified from the Mariana Trench via the culture-dependent method and 16S rRNA sequencing approach. The antimicrobial activity of Microbacterium sp. B1075 was found to be the most potent, and therefore, it was selected as the target strain. Molecular networking analysis via the Global Natural Products Social Molecular Networking (GNPS) platform identified 25 flavonoid compounds as flavonoid secondary metabolites. Among these, genistein was purified and identified as a bioactive compound with significant antibacterial activity. The complete synthesis pathway for genistein was proposed within strain B1075 based on whole-genome sequencing data, with the key gene being CHS (encoding chalcone synthase). The expression of the gene CHS was significantly regulated by high hydrostatic pressure, with a consequent impact on the production of flavonoid compounds in strain B1075, revealing the relationship between actinomycetes' synthesis of flavonoid-like secondary metabolites and their adaptation to high-pressure environments at the molecular level. These results not only expand our understanding of deep-sea microorganisms but also hold promise for providing valuable insights into the development of novel pharmaceuticals in the field of biopharmaceuticals.
Keywords: biological activity; biosynthetic genes; deep-sea actinomycetes; flavonoid compounds.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Ribeiro I., Antunes J.T., Alexandrino D.A.M., Tomasino M.P., Almeida E., Hilário A., Urbatzka R., Leão P.N., Mucha A.P., Carvalho M.F. Actinobacteria from Arctic and Atlantic deep-sea sediments—Biodiversity and bioactive potential. Front. Microbiol. 2023;14:1158441. doi: 10.3389/fmicb.2023.1158441. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

