Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption
- PMID: 38922012
- PMCID: PMC11209563
- DOI: 10.3390/vetsci11060265
Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption
Abstract
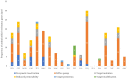
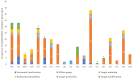
Raw milk and dairy products can serve as potential vectors for transmissible bacterial, viral and protozoal diseases, alongside harboring antimicrobial-resistance genes. This study monitors the changes in the antimicrobial-resistance gene pool in raw milk and cheese, from farm to consumer, utilizing next-generation sequencing. Five parallel sampling runs were conducted to assess the resistance gene pool, as well as phage or plasmid carriage and potential mobility. In terms of taxonomic composition, in raw milk the Firmicutes phylum made up 41%, while the Proteobacteria phylum accounted for 58%. In fresh cheese, this ratio shifted to 93% Firmicutes and 7% Proteobacteria. In matured cheese, the composition was 79% Firmicutes and 21% Proteobacteria. In total, 112 antimicrobial-resistance genes were identified. While a notable reduction in the resistance gene pool was observed in the freshly made raw cheese compared to the raw milk samples, a significant growth in the resistance gene pool occurred after one month of maturation, surpassing the initial gene frequency. Notably, the presence of extended-spectrum beta-lactamase (ESBL) genes, such as OXA-662 (100% coverage, 99.3% identity) and OXA-309 (97.1% coverage, 96.2% identity), raised concerns; these genes have a major public health relevance. In total, nineteen such genes belonging to nine gene families (ACT, CMY, EC, ORN, OXA, OXY, PLA, RAHN, TER) have been identified. The largest number of resistance genes were identified against fluoroquinolone drugs, which determined efflux pumps predominantly. Our findings underscore the importance of monitoring gene pool variations throughout the product pathway and the potential for horizontal gene transfer in raw products. We advocate the adoption of a new approach to food safety investigations, incorporating next-generation sequencing techniques.
Keywords: ESBL; NGS; antimicrobial-resistance genes; cattle; dairy products; food safety; next-generation sequencing; public health; raw milk; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Short communication: Heat-resistant Escherichia coli as potential persistent reservoir of extended-spectrum β-lactamases and Shiga toxin-encoding phages in dairy.J Dairy Sci. 2016 Nov;99(11):8622-8632. doi: 10.3168/jds.2016-11076. Epub 2016 Aug 24. J Dairy Sci. 2016. PMID: 27568050
-
Phenotypic and genotypic characterization of antimicrobial resistances reveals the effect of the production chain in reducing resistant lactic acid bacteria in an artisanal raw ewe milk PDO cheese.Food Res Int. 2024 Jul;187:114308. doi: 10.1016/j.foodres.2024.114308. Epub 2024 Apr 18. Food Res Int. 2024. PMID: 38763625
-
Occurrence and Characteristics of Extended-Spectrum β-Lactamase-Producing Escherichia coli from Dairy Cattle, Milk, and Farm Environments in Peninsular Malaysia.Pathogens. 2020 Nov 30;9(12):1007. doi: 10.3390/pathogens9121007. Pathogens. 2020. PMID: 33266299 Free PMC article.
-
Antibiotic Resistance in Fermented Foods Chain: Evaluating the Risks of Emergence of Enterococci as an Emerging Pathogen in Raw Milk Cheese.Int J Microbiol. 2024 Dec 26;2024:2409270. doi: 10.1155/ijm/2409270. eCollection 2024. Int J Microbiol. 2024. PMID: 39749146 Free PMC article. Review.
-
Graduate Student Literature Review: Current understanding of the influence of on-farm factors on bovine raw milk and its suitability for cheesemaking.J Dairy Sci. 2021 Nov;104(11):12173-12183. doi: 10.3168/jds.2021-20146. Epub 2021 Aug 26. J Dairy Sci. 2021. PMID: 34454752 Review.
Cited by
-
Role of Lysogenic Phages in the Dissemination of Antibiotic Resistance Genes Applied in the Food Chain.Foods. 2025 Mar 21;14(7):1082. doi: 10.3390/foods14071082. Foods. 2025. PMID: 40238219 Free PMC article. Review.
-
Diversity and antibiotic resistance of cultivable bacteria in bulk tank milk from dairy farms in Shandong Province, China.Front Vet Sci. 2025 Jul 23;12:1649876. doi: 10.3389/fvets.2025.1649876. eCollection 2025. Front Vet Sci. 2025. PMID: 40771950 Free PMC article.
References
-
- Benmazouz I., Kövér L., Kardos G. The Rise of Antimicrobial Resistance in Wild Birds: Potential AMR Sources and Wild Birds as AMR Reservoirs and Disseminators: Literature Review. Hung. Vet. J. 2024;146:91–105. doi: 10.56385/magyallorv.2024.02.91-105. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

