Allosteric Effects of EF-G Domain I Mutations Inducing Ribosome Frameshifting Revealed by Multiplexed Force Spectroscopy
- PMID: 38923096
- PMCID: PMC11446648
- DOI: 10.1002/cbic.202400130
Allosteric Effects of EF-G Domain I Mutations Inducing Ribosome Frameshifting Revealed by Multiplexed Force Spectroscopy
Abstract
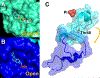
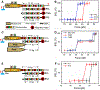
Ribosome translocation catalyzed by elongation factor G (EF-G) is a critical step in protein synthesis where the ribosome typically moves along the mRNA by three nucleotides at each step. To investigate the mechanism of EF-G catalysis, it is essential to precisely resolve the ribosome motion at both ends of the mRNA, which, to our best knowledge, is only achieved with the magnetic-based force spectroscopy developed by our groups. Here, we introduce a novel multiplexed force spectroscopy technique that, for the first time, offers single-nucleotide resolution for multiple samples. This technique combines multiple acoustic force generators with the smallest atomic magnetometer designed for biological research. Utilizing this technique, we demonstrate that mutating EF-G at the GTP binding pocket results in the ribosome moving only two nucleotides on both ends of the mRNA, thereby compromising ribosome translocation. This finding suggests a direct link between GTP hydrolysis and ribosome translocation. Our results not only provide mechanistic insights into the role of GTP binding pocket but also illuminate how allosteric mutations can manipulate translocation. We anticipate broader applications of our technique in the ribosome field, leveraging its high efficiency and single-nucleotide resolution.
Keywords: Allosteric effect; DNA ruler; Elongation factor G mutation; Frameshifting; Multiplexed force spectroscopy; Ribosome translocation.
© 2024 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of Interests
The authors declare no competing interests.
Figures
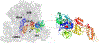

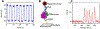




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

