CK2 phosphorylation of CMTR1 promotes RNA cap formation and influenza virus infection
- PMID: 38923463
- PMCID: PMC11290353
- DOI: 10.1016/j.celrep.2024.114405
CK2 phosphorylation of CMTR1 promotes RNA cap formation and influenza virus infection
Abstract
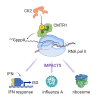
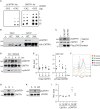
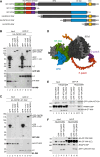
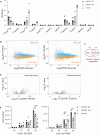
The RNA cap methyltransferase CMTR1 methylates the first transcribed nucleotide of RNA polymerase II transcripts, impacting gene expression mechanisms, including during innate immune responses. Using mass spectrometry, we identify a multiply phosphorylated region of CMTR1 (phospho-patch [P-Patch]), which is a substrate for the kinase CK2 (casein kinase II). CMTR1 phosphorylation alters intramolecular interactions, increases recruitment to RNA polymerase II, and promotes RNA cap methylation. P-Patch phosphorylation occurs during the G1 phase of the cell cycle, recruiting CMTR1 to RNA polymerase II during a period of rapid transcription and RNA cap formation. CMTR1 phosphorylation is required for the expression of specific RNAs, including ribosomal protein gene transcripts, and promotes cell proliferation. CMTR1 phosphorylation is also required for interferon-stimulated gene expression. The cap-snatching virus, influenza A, utilizes host CMTR1 phosphorylation to produce the caps required for virus production and infection. We present an RNA cap methylation control mechanism whereby CK2 controls CMTR1, enhancing co-transcriptional capping.
Keywords: CMTR1; CP: Microbiology; CP: Molecular biology; RNA; RNA cap; cell proliferation; influenza virus; innate immunity; ribosomes; transcription; translation.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

