An ANXA11 P93S variant dysregulates TDP-43 and causes corticobasal syndrome
- PMID: 38923692
- PMCID: PMC11350008
- DOI: 10.1002/alz.13915
An ANXA11 P93S variant dysregulates TDP-43 and causes corticobasal syndrome
Abstract
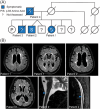
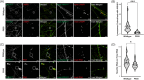
Introduction: Variants of uncertain significance (VUS) surged with affordable genetic testing, posing challenges for determining pathogenicity. We examine the pathogenicity of a novel VUS P93S in Annexin A11 (ANXA11) - an amyotrophic lateral sclerosis/frontotemporal dementia-associated gene - in a corticobasal syndrome kindred. Established ANXA11 mutations cause ANXA11 aggregation, altered lysosomal-RNA granule co-trafficking, and transactive response DNA binding protein of 43 kDa (TDP-43) mis-localization.
Methods: We described the clinical presentation and explored the phenotypic diversity of ANXA11 variants. P93S's effect on ANXA11 function and TDP-43 biology was characterized in induced pluripotent stem cell-derived neurons alongside multiomic neuronal and microglial profiling.
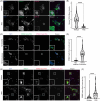
Results: ANXA11 mutations were linked to corticobasal syndrome cases. P93S led to decreased lysosome colocalization, neuritic RNA, and nuclear TDP-43 with cryptic exon expression. Multiomic microglial signatures implicated immune dysregulation and interferon signaling pathways.
Discussion: This study establishes ANXA11 P93S pathogenicity, broadens the phenotypic spectrum of ANXA11 mutations, underscores neuronal and microglial dysfunction in ANXA11 pathophysiology, and demonstrates the potential of cellular models to determine variant pathogenicity.
Highlights: ANXA11 P93S is a pathogenic variant. Corticobasal syndrome is part of the ANXA11 phenotypic spectrum. Hybridization chain reaction fluorescence in situ hybridization (HCR FISH) is a new tool for the detection of cryptic exons due to TDP-43-related loss of splicing regulation. Microglial ANXA11 and related immune pathways are important drivers of disease. Cellular models are powerful tools for adjudicating variants of uncertain significance.
Keywords: ANXA11; TDP‐43; corticobasal syndrome; variant of uncertain significance.
© 2024 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association. This article has been contributed to by U.S. Government employees and their work is in the public domain in the USA.
Conflict of interest statement
Jennifer S. Yokoyama serves on the scientific advisory board for the Epstein Family Alzheimer's Research Collaboration. Debora S. Marks is an advisor for Dyno Therapeutics, Octant, Jura Bio, Tectonic Therapeutic, and Genentech and is a co‐founder of Seismic Therapeutic. The authors Allison Snyder, Veronica H. Ryan, James Hawrot, Sydney Lawton, Daniel M. Ramos, Y. Andy Qi, Kory Johnson, Xylena Reed, Nicholas L. Johnson, Aaron W. Kollasch, Megan Duffy, Lawren VandeVrede, J. Nicholas Cochran, Camilo Toro, Bibiana Bielekova, Justin Y. Kwan, Mark R. Cookson, and Michael E. Ward report no competing interests. Author disclosures are available in the supporting information.
Figures



Update of
-
An ANXA11 P93S variant dysregulates TDP-43 and causes corticobasal syndrome.Res Sq [Preprint]. 2023 Oct 19:rs.3.rs-3462973. doi: 10.21203/rs.3.rs-3462973/v1. Res Sq. 2023. Update in: Alzheimers Dement. 2024 Aug;20(8):5220-5235. doi: 10.1002/alz.13915. PMID: 37886540 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- K01AG049152/NIH National Institute on Aging
- 75N95022C00031/DA/NIDA NIH HHS/United States
- K01 AG049152/AG/NIA NIH HHS/United States
- U54NS123985/NIH National Institute of Neurological Disorders and Stroke
- ZIAAG000534/AG/NIA NIH HHS/United States
- FI2 GM142475/GM/NIGMS NIH HHS/United States
- U54 NS123985/NS/NINDS NIH HHS/United States
- P30AG062422/NIH National Institute on Aging
- 1ZIAAG000539-01/NIH National Institute on Aging
- R00 AG068271/AG/NIA NIH HHS/United States
- Global Brain Health Institute
- NIH Undiagnosed Diseases Program, Undiagnosed Diseases Network
- P50 AG023501/AG/NIA NIH HHS/United States
- National Institute of Allergy and Infectious Diseases
- R01 AG062588/AG/NIA NIH HHS/United States
- ALZ/Alzheimer's Association/United States
- R00AG068271/NIH National Institute on Aging
- P01AG019724/NIH National Institute on Aging
- U19 AG079774/AG/NIA NIH HHS/United States
- R01AG057234/NIH National Institute on Aging
- FI2GM142475/NIH National Institute of General Medical Sciences
- P30 AG062422/AG/NIA NIH HHS/United States
- K23AG073514/NIH National Institute on Aging
- P01 AG019724/AG/NIA NIH HHS/United States
- 2016-A-005-SUP/Larry L. Hillblom Foundation
- NIH Office of Intramural Training and Education
- R01 AG057234/AG/NIA NIH HHS/United States
- ZIA AG000534/ImNIH/Intramural NIH HHS/United States
- Chan-Zuckerberg Initiative's Neurodegeneration Challenge Network
- ZIA AG000539/ImNIH/Intramural NIH HHS/United States
- Rainwater Charitable Foundation
- Bluefield Project to Cure Frontotemporal Dementia
- French Foundation
- U19AG079774/NIH National Institute on Aging
- P50AG023501/NIH National Institute on Aging
- Intramural Research Programs of the National Institute of Neurological Disorders and Stroke
- P01AG1972403/NIH National Institute on Aging
- HudsonAlpha Foundation Memory and Mobility Fund
- R01AG062588/NIH National Institute on Aging
- K23 AG073514/AG/NIA NIH HHS/United States
- Mary Oakley Foundation
LinkOut - more resources
Full Text Sources
Miscellaneous

