Measurement of protein concentration in bacteria and small organelles under a light transmission microscope
- PMID: 38923720
- PMCID: PMC11323175
- DOI: 10.1002/jmr.3099
Measurement of protein concentration in bacteria and small organelles under a light transmission microscope
Abstract
Protein concentration (PC) is an essential characteristic of cells and organelles; it determines the extent of macromolecular crowding effects and serves as a sensitive indicator of cellular health. A simple and direct way to quantify PC is provided by brightfield-based transport-of-intensity equation (TIE) imaging combined with volume measurements. However, since TIE is based on geometric optics, its applicability to micrometer-sized particles is not clear. Here, we show that TIE can be used on particles with sizes comparable to the wavelength. At the same time, we introduce a new ImageJ plugin that allows TIE image processing without resorting to advanced mathematical programs. To convert TIE data to PC, knowledge of particle volumes is essential. The volumes of bacteria or other isolated particles can be measured by displacement of an external absorbing dye ("transmission-through-dye" or TTD microscopy), and for spherical intracellular particles, volumes can be estimated from their diameters. We illustrate the use of TIE on Escherichia coli, mammalian nucleoli, and nucleolar fibrillar centers. The method is easy to use and achieves high spatial resolution.
Keywords: bacteria; cell volume; fibrillar centers; macromolecular crowding; nucleoli; refractive index; transport‐of‐intensity equation.
© 2024 The Author(s). Journal of Molecular Recognition published by John Wiley & Sons Ltd.
Conflict of interest statement
Conflict of Interest
The authors declare that they have no conflict of interest.
Figures

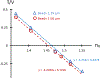
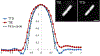
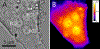
References
-
- Mittal S, Chowhan RK, Singh LR. Macromolecular crowding: Macromolecules friend or foe. Biochim Biophys Acta Gen Subj 1850:1822–1831 (2015). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

