"Find Me" and "Eat Me" signals: tools to drive phagocytic processes for modulating antitumor immunity
- PMID: 38923737
- PMCID: PMC11260773
- DOI: 10.1002/cac2.12579
"Find Me" and "Eat Me" signals: tools to drive phagocytic processes for modulating antitumor immunity
Abstract
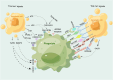
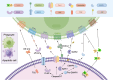
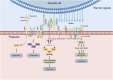
Phagocytosis, a vital defense mechanism, involves the recognition and elimination of foreign substances by cells. Phagocytes, such as neutrophils and macrophages, rapidly respond to invaders; macrophages are especially important in later stages of the immune response. They detect "find me" signals to locate apoptotic cells and migrate toward them. Apoptotic cells then send "eat me" signals that are recognized by phagocytes via specific receptors. "Find me" and "eat me" signals can be strategically harnessed to modulate antitumor immunity in support of cancer therapy. These signals, such as calreticulin and phosphatidylserine, mediate potent pro-phagocytic effects, thereby promoting the engulfment of dying cells or their remnants by macrophages, neutrophils, and dendritic cells and inducing tumor cell death. This review summarizes the phagocytic "find me" and "eat me" signals, including their concepts, signaling mechanisms, involved ligands, and functions. Furthermore, we delineate the relationships between "find me" and "eat me" signaling molecules and tumors, especially the roles of these molecules in tumor initiation, progression, diagnosis, and patient prognosis. The interplay of these signals with tumor biology is elucidated, and specific approaches to modulate "find me" and "eat me" signals and enhance antitumor immunity are explored. Additionally, novel therapeutic strategies that combine "find me" and "eat me" signals to better bridge innate and adaptive immunity in the treatment of cancer patients are discussed.
Keywords: CARL; CX3CL1; Fc; LPC; Phagocytosis; PtSer; SLAMF7; cancer immunotherapy; “Eat me” signal; “Find me” signal.
© 2024 The Author(s). Cancer Communications published by John Wiley & Sons Australia, Ltd on behalf of Sun Yat‐sen University Cancer Center.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
- SYS202202/Shandong Provincial Laboratory Project
- 81972888/National Natural Science Foundation of China
- 82272819/National Natural Science Foundation of China
- JNL-202219B/Research Project of Jinan Microecological Biomedicine Shandong Laboratory
- JNL-202204A/Research Project of Jinan Microecological Biomedicine Shandong Laboratory
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

